Definition and Biological Significance
SUMOylation, a reversible post-translational modification, has garnered substantial recognition for its critical role in various cellular functions. This modification is ubiquitous across all eukaryotic organisms, serving essential roles in maintaining genomic stability, regulating transcription, controlling gene expression, and modulating intracellular signal transduction pathways. Analogous to ubiquitination, SUMOylation involves the covalent attachment of small ubiquitin-like modifier (SUMO) proteins to the lysine residues of substrate proteins. By modulating the biological activity of these substrates, SUMOylation exerts significant influence on both physiological and pathological cellular processes, with aberrant activation notably observed in the development and progression of various malignancies.
Mechanism and Functional Implications of SUMOylation
In contrast to ubiquitination, SUMOylation does not promote the degradation of target proteins but rather contributes to their stabilization. As a reversible process, the catalytic cycle of SUMOylation encompasses several steps, including maturation, activation, conjugation, ligation, and de-modification. This cycle involves the participation of E1 (Aos1/Uba2), E2 (Ubc9), multiple E3 ligases, and SUMO-specific proteases such as SENPs (sentrin/SUMO-specific proteases). To date, four isoforms of SUMO have been identified: SUMO1, SUMO2, SUMO3, and SUMO4. This paper will provide a concise overview of the SUMOylation cycle and its functional implications.
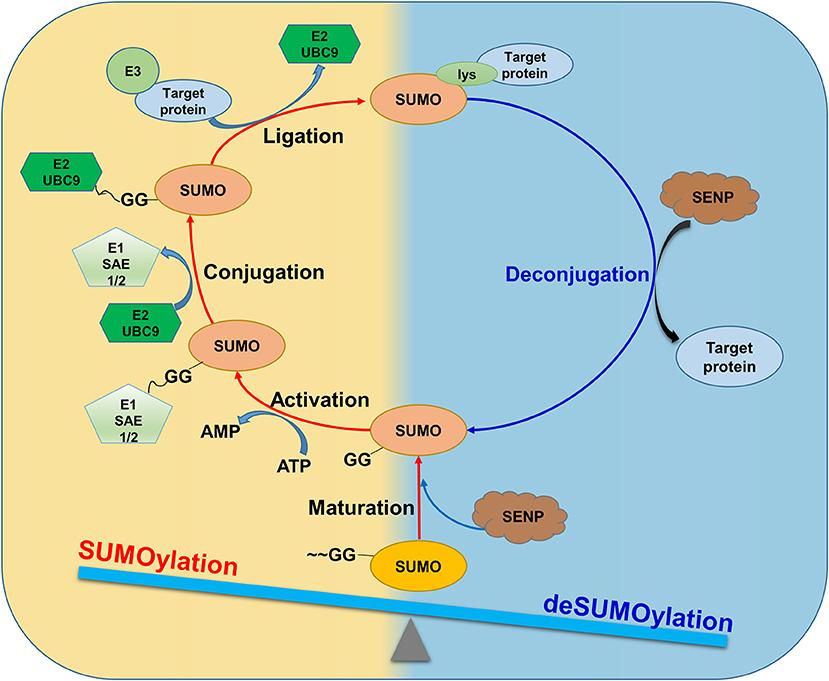 Figure 1. SUMOylation and deSUMOylation are dynamically balanced under physiological conditions. SUMO proteins are proteolytically cleaved by SUMO/ SENP to form mature forms.
Figure 1. SUMOylation and deSUMOylation are dynamically balanced under physiological conditions. SUMO proteins are proteolytically cleaved by SUMO/ SENP to form mature forms.
SUMO Proteins and the SUMOylation Catalytic Cycle
SUMO proteins, which share significant structural homology with ubiquitin, are highly conserved throughout evolution and are present in protozoa, metazoans, plants, and fungi. Although the SUMOylation cycle bears resemblance to the ubiquitination cycle in terms of the processes involved, the functional outcomes of these modifications are distinctly different. The primary function of ubiquitination is to tag target proteins for recognition and degradation by the proteasome, whereas SUMOylation tends to enhance the stability of target proteins.
SUMOylation facilitates the fine-tuning of protein-protein interactions, thereby mediating the localization and functional regulation of substrate proteins. The catalytic mechanism of SUMOylation, which is consistent across all SUMO proteins, encompasses several key steps: maturation, activation, conjugation, ligation, and de-modification (Figure 2). These steps collectively constitute the complete SUMOylation cycle, ensuring the precise modulation of substrate proteins.
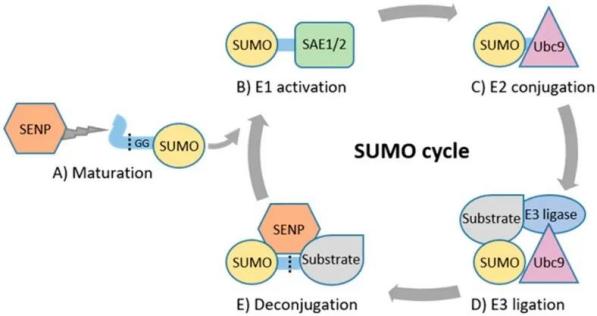 Figure 2. Catalytic Cycle of SUMOylation
Figure 2. Catalytic Cycle of SUMOylation
Maturation:
The SUMO proteins, with a molecular weight of approximately 11 kDa, initially exist as inactive precursor molecules. These precursors are processed by members of the SUMO1/sentrin-specific protease (SENP) family, which cleave four amino acids from the C-terminus, exposing a terminal diglycine (GG) motif.
Activation:
The SUMO E1 activating enzyme is a 110 kDa protein comprising two subunits, SAE1 (SUMO-activating enzyme 1) or Aos1 and SAE2 (SUMO-activating enzyme 2) or Uba2. These subunits typically form a heterodimer, SAE1-SAE2 or Aos1-Uba2. The heterodimer facilitates the ATP-dependent formation of a SUMO-adenylate intermediate. Subsequently, the C-terminal carboxyl group of SUMO forms a thioester bond with the cysteine residue of SAE2/Uba2, thereby activating the SUMO protein.
Conjugation:
The sole SUMO-conjugating enzyme E2 (SUMO E2 or Ubc9) is located either on the cytoplasmic side or the nucleoplasmic side of the nuclear pore complex (NPC). The activated SUMO protein is transferred to the conserved cysteine residue at position 93 of Ubc9 via a transesterification reaction, forming an E2-SUMO thioester complex. Ubc9 then conjugates the SUMO molecule to the lysine residue of the target protein through an isopeptide bond, completing the SUMOylation process.
Ligation:
SUMO E3 ligases are crucial for the substrate-specific targeting of SUMO proteins. These ligases enhance the interaction between the substrate and the SUMO-E2 complex, facilitating the transfer of SUMO from Ubc9 to the substrate. By precisely positioning the SUMO-E2 complex, SUMO E3 ligases reduce the distance between the thioester bond of the SUMO-E2 complex and the lysine residue of the substrate, thereby increasing substrate specificity.
De-modification:
SUMOylation is a reversible process wherein de-modification involves the cleavage of the terminal glycine residues of SUMO by specific proteases, removing SUMO from the lysine residues of the target protein. To date, six SENP family proteases have been identified in mammals or humans: SENP1, SENP2, SENP3, SENP5, SENP6, and SENP7. SENP1 and SENP2 can deconjugate SUMO1 and SUMO2/3 proteins, while SENP3 and SENP5 primarily deconjugate SUMO2/3 proteins. SENP6 and SENP7 disassemble SUMO2/3 poly-chains. The removal of SUMO proteins by SENPs frees them for another catalytic cycle.
Biological Functions of SUMOylation
SUMOylation, the covalent attachment of SUMO proteins to target substrates, serves as a rapid and reversible regulatory mechanism influencing a multitude of cellular processes. These processes include gene expression, transcriptional regulation, DNA damage response (DDR), nucleocytoplasmic transport, cell signaling, mRNA maturation, meiosis, mitosis, chromatin remodeling, ion channel activity, cell cycle regulation, and apoptosis.
SUMOylation exerts a substantial impact on various intracellular pathways. In the NFκB signaling pathway, SUMOylation and deSUMOylation of essential modulatory proteins can be observed in both the cytoplasm and nucleus, particularly involving NFκB essential modulator (NEMO). This modification can either positively or negatively regulate NFκB activation, demonstrating its role as a critical modulator.
Within the JAK-STAT signaling pathway, SUMOylation of signal transducer and activator of transcription (STAT) proteins inhibits their phosphorylation, subsequently repressing the expression of growth factor-induced target genes.
Similarly, in the TGFβ signaling pathway, SUMOylation of SMAD proteins modulates the transcription of target genes, exerting both promotive and inhibitory effects depending on the cellular context and specific SMAD involved.
In summary, SUMOylation is a versatile post-translational modification that plays a pivotal role in fine-tuning cellular functions and maintaining cellular homeostasis through its dynamic regulation of key signaling pathways.
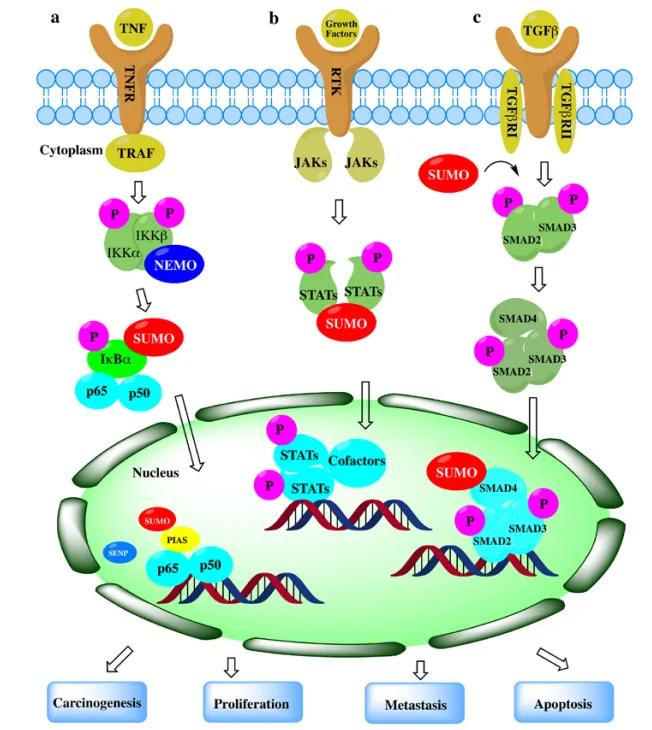 Figure 3. Role of SUMOylation in Representative Cancer Pathways
Figure 3. Role of SUMOylation in Representative Cancer Pathways
SUMOylation, a crucial post-translational modification, plays diverse roles in cellular processes, particularly in transcriptional regulation and chromatin remodeling. Understanding its impact on specific SUMO targets and sites requires meticulous investigation due to significant variations in its effects.
Figure 4 illustrates mechanisms involving transcription factors crucial in cancer development affected by SUMOylation. SUMO (depicted by blue circles, "S") modulates protein interactions, hindering binding with transcriptional repressors and promoting the formation of inhibitory protein complexes. For instance:
SUMOylation of FoxM1 (Panel A) inhibits protein dimerization, thereby inducing transcriptional activity.
SUMOylation of transcription repressor TEL/ETV6 (Panel B) disrupts multimer formation essential for inhibitory promoter region transcriptional activity, thereby enhancing gene expression.
Mutant MITF lacking SUMOylation (Panel C) shows enhanced DNA binding and increased expression of hypoxia-inducible factor 1 (HIF1/), suggesting SUMOylation inhibits DNA binding and reduces transcriptional activity.
SUMOylation of IkB/ (Panel D) stabilizes the protein, blocking gene expression by competing with ubiquitin (depicted by red circles, "U") at the same lysine (K) residues as NFκB inhibitor IkB/. Ubiquitination of IkB/ leads to proteasomal degradation and release of p65 and p50 subunits, enhancing transcriptional activity.
Moreover, multiple SUMO moieties can facilitate ubiquitination via SUMO-targeted ubiquitin ligases (Stubl), potentially affecting c-Myc. SUMOylation of PIAS1 (Panel D) activates Stubl and ring finger protein 4 (RNF4), promoting ubiquitination and subsequent proteasomal degradation, thereby reducing transcriptional activity.
In summary, SUMOylation exerts complex regulatory effects on transcription factors within cancer pathways, highlighting its role in modulating gene expression and cellular responses critical for oncogenesis and tumor progression.
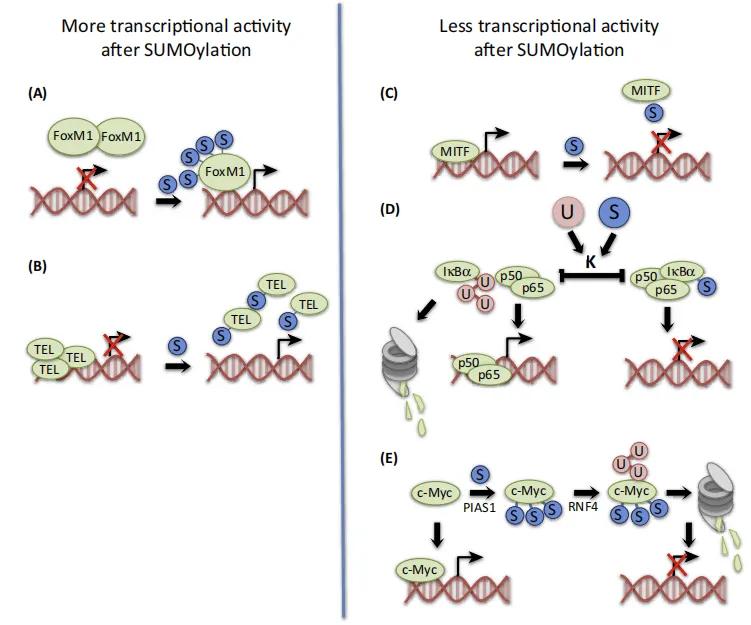 Figure 4. Regulation of Transcription Factor Activity by Small Ubiquitin-like Modifier (SUMO) Acylation
Figure 4. Regulation of Transcription Factor Activity by Small Ubiquitin-like Modifier (SUMO) Acylation
Current research is investigating drugs targeting the SUMO-activating enzyme. Chemical inhibitors, such as ginkgolic acid and perillaldehyde, bind to the SUMO-activating enzyme and obstruct the formation of the E1-SUMO intermediate. Several chemotherapeutic agents, used in the treatment of acute myeloid leukemia, have been reported to induce the formation of reactive oxygen species (ROS), subsequently inhibiting the SUMO pathway and reducing tumor growth.
Despite the potential of targeting the SUMO pathway for cancer therapy, the development of specific small molecule inhibitors that selectively inhibit only the enzymes of interest remains challenging. This necessitates a deeper understanding of the regulatory pathways and exact mechanisms of the SUMO system, as well as the SUMOylation signals that trigger specific SUMO targets involved in cancer progression.
Research Directions in SUMOylation
Functional Relevance of Novel SUMOylated Substrates:
What are the functional implications of the recently discovered novel SUMOylated substrates?
Using proteomics approaches, numerous new SUMO target proteins have been identified in a site-specific manner. These targets include critical cell cycle regulators.
Substrate Specificity of SUMOylating and DeSUMOylating Enzymes:
What are the specific SUMO target protein subgroups for each SUMOylating and deSUMOylating enzyme?
The SUMOylation and deSUMOylation mechanisms involve a relatively limited number of enzymes. Given the complexity of the SUMO proteome, each SUMO E3 ligase and SUMO protease must regulate a relatively large number of SUMO target proteins.
Cooperative Role of Post-Translational Modifications in Cell Cycle Progression:
How do different post-translational modifications cooperate in a holistic cellular context to drive cell cycle progression?
Similar to SUMOylation, ubiquitination, phosphorylation, and other post-translational modifications play crucial roles in cell cycle progression.
Mechanisms of SUMO Redistribution During the Cell Cycle:
What are the mechanisms underlying the redistribution of the SUMO machinery during cell cycle progression?
Various cancer types rely on a functional SUMOylation system. Is it feasible to develop SUMOylation and deSUMOylation inhibitors for cancer therapy? Can healthy cells tolerate these inhibitors?
Select Service
Detection Methods for Protein SUMOylation
Immunoprecipitation (Co-IP) Followed by Western Blot (WB) Analysis for SUMO Proteins
To detect SUMOylated proteins, cell lysis is performed using lysis buffer S, which comprises 20 mM sodium phosphate (pH 7.4), 150 mM NaCl, 1% SDS, 1% Triton X-100, 0.5% sodium deoxycholate, 5 mM EDTA, 5 mM EGTA, 20 mM N-ethylmaleimide (NEM), 5 mM dithiothreitol (DTT), and a protease inhibitor cocktail. The cell lysate is then subjected to boiling for 10 minutes, followed by sonication until the lysate becomes liquid. The lysate is clarified by centrifugation at 16,000g for 10 minutes at 4°C. The supernatant is subsequently diluted 1:10 with lysis buffer S devoid of SDS. The diluted lysate is incubated overnight at 4°C with anti-SUMO or anti-target protein antibodies conjugated to protein A/G magnetic beads. The beads are washed five times with high-salt buffer and then boiled in SDS loading buffer for 10 minutes. The samples are analyzed by WB for SUMO1/2/3 or the target protein. The results can be referenced from the figure below:
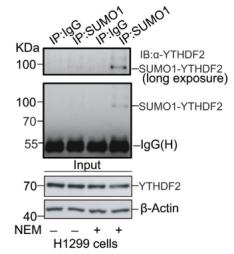 Figure 5. WB results
Figure 5. WB results
GST Pull-Down Assay for Detecting Protein SUMOylation
To detect the SUMOylation of target proteins, the recombinant target protein tagged with GST (glutathione S-transferase) is expressed in BL21 (DE3) Escherichia coli cells. Transformation is performed with the GST-tagged recombinant target protein plasmid alone or co-transformed with pE1E2S1. Protein expression is induced with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at 16°C for 12 hours. The cells are lysed in PBS-L buffer containing 50 mM NaH2PO4 (pH 7.2), 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM dithiothreitol (DTT), 1 mM EDTA, and 0.1% (v/v) Triton X-100. The lysate is sonicated on ice for 10 minutes and subsequently centrifuged at 17,000g for 20 minutes. The supernatant is incubated with glutathione resin at 4°C for 4 hours. The resin is washed three times with PBS-L buffer and eluted with GSH buffer comprising 50 mM Tris (pH 8.0) and 20 mM reduced glutathione (GSH). The purified protein is analyzed by Western blotting for SUMO1/2/3. The results can be referenced from the figure below:
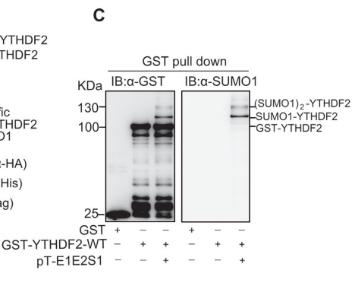 Figure 6. GST pull down results
Figure 6. GST pull down results
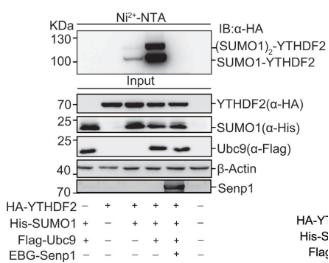
Analysis of SUMO Proteins Using Ni²⁺-NTA Magnetic Beads
To analyze SUMOylated proteins, cells are harvested 48 hours post-transfection with a His-SUMO construct. Approximately 25% of the collected cells are subjected to lysis using M-RIPA buffer and analyzed by Western blotting. The remaining cells are lysed in 3 mL of His lysis buffer. Subsequently, 60 μL of Ni²⁺-NTA agarose beads are added to the lysate, which is incubated with rotation at 4°C overnight. The beads are sequentially washed at room temperature with 750 μL of each washing buffer for 5 minutes. The washing buffers used are as follows: Washing Buffer 1, Washing Buffer 2, Washing Buffer 3, and Washing Buffer 4. After the final wash, His6-tagged SUMO-conjugated target proteins are eluted with 75 μL of elution buffer for 20 minutes and analyzed by Western blotting.
- Hydrolysis Buffer: 6 M guanidine hydrochloride, 0.1 M Na2HPO4/NaH2PO4, 0.01 M Tris/HCl (pH 8.0), 5 mM imidazole, 10 mM β-mercaptoethanol.
- Washing Buffer 1: 6 M guanidine hydrochloride, 0.1 M Na2HPO4/NaH2PO4, 0.01 M Tris/HCl (pH 8.0), 10 mM β-mercaptoethanol.
- Washing Buffer 2: 8 M urea, 0.1 M Na2HPO4/NaH2PO4, 0.01 M Tris/HCl (pH 8.0), 10 mM β-mercaptoethanol.
- Washing Buffer 3: 8 M urea, 0.1 M Na2HPO4/NaH2PO4, 0.01 M Tris/HCl (pH 6.3), 10 mM β-mercaptoethanol (Buffer A) with 0.2% Triton X-100.
- Washing Buffer 4: Buffer A with 0.1% Triton X-100.
- Elution Buffer: 200 mM imidazole, 0.15 M Tris/HCl (pH 6.7), 30% glycerol, 0.72 M β-mercaptoethanol, 5% SDS.
The results can be referenced from the figure below:
References
- Chang Hui-Ming,Yeh Edward T H,SUMO: From Bench to Bedside. Physiol Rev, 2020, 100: 1599-1619.
- Fan Yao,Li Xiang,Zhang Lei et al. SUMOylation in Viral Replication and Antiviral Defense. Adv Sci (Weinh), 2022, 9: e2104126.
- Hertz Emil P T,Vega Ignacio Alonso-de,Kruse Thomas et al. The SUMO-NIP45 pathway processes toxic DNA catenanes to prevent mitotic failure. Nat Struct Mol Biol, 2023, 30: 1303-1313.
- Tan Dan,Lu Meiping,Cai Yuqing et al. SUMOylation of Rho-associated protein kinase 2 induces goblet cell metaplasia in allergic airways. Nat Commun, 2023, 14: 3887.
- Mo Yongzhen,Wang Yumin,Zhang Shuai et al. Circular RNA circRNF13 inhibits proliferation and metastasis of nasopharyngeal carcinoma via SUMO2. Mol Cancer, 2021, 20: 112.
- Hou, G., Zhao, X., Li, L., Yang, Q., Liu, X., Huang, C., Lu, R., Chen, R., Wang, Y., Jiang, B., & Yu, J. (2021). SUMOylation of YTHDF2 promotes mRNA degradation and cancer progression by increasing its binding affinity with m6A-modified mRNAs. Nucleic acids research, 49(5), 2859–2877.
- Fox, B.M.; Janssen, A.; Estevez-Ordonez, D.; Gessler, F.; Vicario, N.; Chagoya, G.; Elsayed, G.; Sotoudeh, H.; Stetler, W.; Friedman, G.K.; et al. SUMOylation in Glioblastoma: A Novel Therapeutic Target. Int. J. Mol. Sci. 2019
- Zhao W, Zhang X, Rong J. SUMOylation as a Therapeutic Target for Myocardial Infarction. Front Cardiovasc Med. 2021
Our products and services are for research use only.