Protein post-translational modifications (PTMs) refer to the covalent addition or removal of specific functional groups to amino acids within proteins. PTMs play a critical role in regulating protein activity, stability, expression, localization, and interactions with other proteins, thus influencing nearly all biochemical processes within the cell. Common PTMs include ubiquitination, phosphorylation, acetylation, glycosylation, and SUMOylation.
What is PTM Crosstalk
The interplay between various types of PTMs, known as "PTM crosstalk," involves intricate interconnections where these modifications influence each other through antagonistic or synergistic actions, collectively regulating biological activities in plants (Venne et al., 2014). PTM crosstalk can occur via modifying enzymes or substrate proteins.
Enzyme-Centric PTM Crosstalk
Enzyme-centric PTM crosstalk involves enzymes that catalyze different PTMs modifying each other, thereby regulating enzymatic activities. In this scenario, one PTM enzyme becomes the substrate for another PTM enzyme.
Substrate-Centric PTM Crosstalk
Substrate-centric PTM crosstalk involves different types of modifications targeting the same protein. This can be further categorized into two situations: (1) different modifications occurring at the same amino acid residue on a single protein, and (2) different modifications occurring at different amino acid residues on the same protein. Hunter (2007) classified PTM crosstalk into positive and negative types. Positive crosstalk occurs when an initial PTM serves as a trigger for the addition or removal of a second PTM, or as a recognition site for other proteins. Conversely, negative crosstalk includes direct competition between two PTMs for the same amino acid residue, or when one PTM obscures the recognition site of another, preventing its addition or removal (Figure 1).
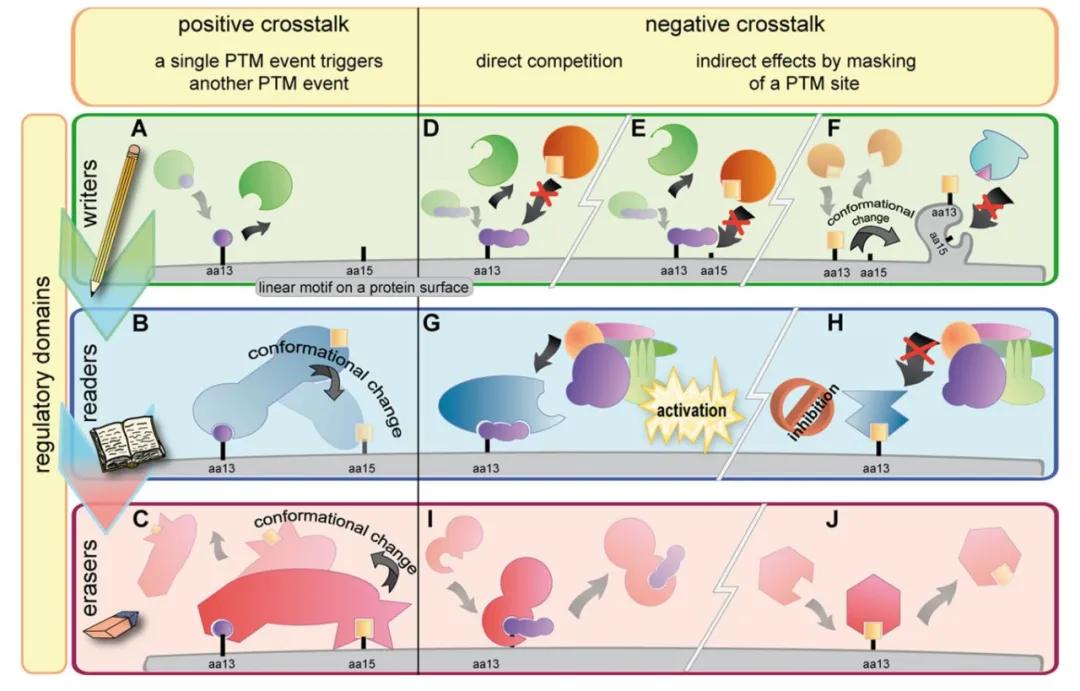 Figure 1 illustrates the classification of PTM crosstalk (Venne et al., 2014). On the surface of proteins, short linear motifs play pivotal roles, facilitating protein-protein interactions and being influenced by distinct regulatory domains. These motifs are distinctly categorized based on their functional properties as writers, readers, and erasers, depicted in green, blue, and red, respectively.
Figure 1 illustrates the classification of PTM crosstalk (Venne et al., 2014). On the surface of proteins, short linear motifs play pivotal roles, facilitating protein-protein interactions and being influenced by distinct regulatory domains. These motifs are distinctly categorized based on their functional properties as writers, readers, and erasers, depicted in green, blue, and red, respectively.
(A) PTMs are precisely attached to specific amino acids (aa) within target proteins.
(B) This initial PTM can attract and trigger the addition of a second PTM, achieved either by recruiting another writer enzyme or through conformational changes induced in reader proteins.
(C) Conversely, PTMs can be recognized and removed by eraser proteins.
(D) In some instances, two distinct PTMs may compete for the same residue attachment site.
(E) Although two PTMs may have different binding sites, the attachment of the first PTM may indirectly obscure the second binding site.
(F) The first PTM might induce conformational changes in the substrate protein, rendering the binding site for the second PTM inaccessible to its writer enzyme.
(G) The attachment of PTMs can initiate various downstream events: upon PTM addition to a target protein, protein complexes may form, triggering specific biochemical pathways.
(H) Another PTM might inhibit the activation of such pathways by blocking the corresponding binding site.
(I, J) In certain scenarios, attached PTMs can also be removed.
Select Service
Types of Ubiquitination-Centric Modification Crosstalk
ubiquitination is one of the most prevalent PTMs in eukaryotic cells and serves as a crucial regulator of plant immune responses (Zhou and Zeng, 2016). The crosstalk between ubiquitination and other PTMs has been extensively documented in plants (Zhao et al., 2014), encompassing interactions with phosphorylation, SUMOylation, poly(ADP-ribosyl)ation, acetylation, glycosylation, and redox modifications (Figure 2) (Zhang and Zeng, 2020).
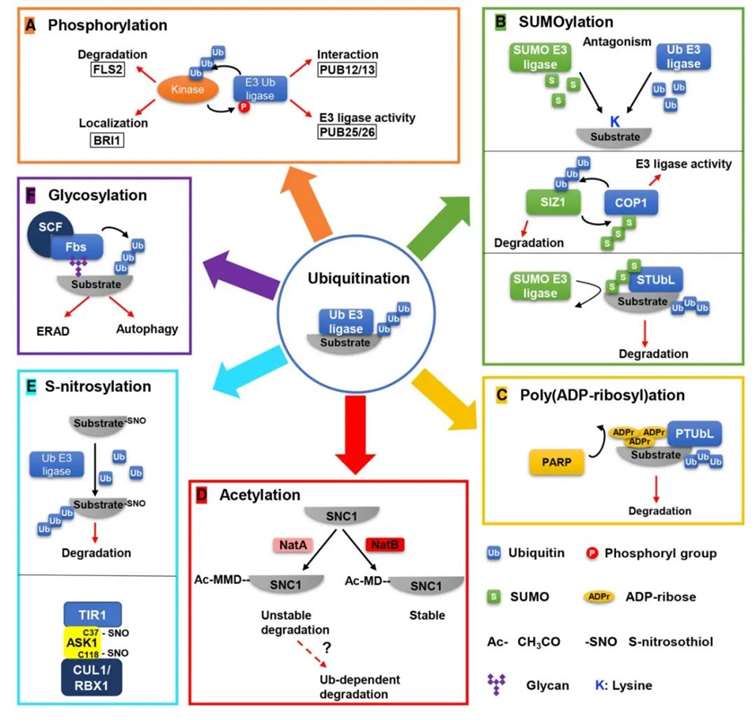 Figure 2: Examples of crosstalk between ubiquitination and other PTMs (Zhang and Zeng, 2020).
Figure 2: Examples of crosstalk between ubiquitination and other PTMs (Zhang and Zeng, 2020).
(A) Ubiquitination and Phosphorylation Crosstalk: Ubiquitination of kinases leads to their degradation (e.g., FLS2) or alters their subcellular localization (e.g., BRI1). Phosphorylation of E3 ligases affects their interaction with substrates (e.g., PUB12/13) or enzymatic activity (e.g., PUB25/26).
(b) Ubiquitination and SUMOylation Crosstalk: Ubiquitination and SUMOylation can competitively target the same lysine residue on substrates. SUMOylation of the E3 ligase COP1 enhances its trans-ubiquitination activity, while ubiquitination of the SUMO E3 ligase SIZ1 mediates its degradation. SUMO-targeted ubiquitin ligases (STUbLs) specifically ubiquitinate and degrade SUMOylated substrates.
(C) Ubiquitination and Poly(ADP-ribosyl)ation Crosstalk: Poly(ADP-ribose)-targeted ubiquitin ligases (PTUbLs) specifically ubiquitinate and degrade poly(ADP-ribosyl)ated substrates.
(D) Ubiquitination and Acetylation Crosstalk: The N-terminal acetylation (NTA) of the first Met (Ac-MMD) in the SNC1 sequence by NatA serves as a degradation signal, potentially dependent on ubiquitination.
(E) Ubiquitination and S-nitrosylation Crosstalk: S-nitrosylation of substrates promotes their ubiquitination and subsequent degradation. S-nitrosylation of Cys37 and Cys118 in ASK1 is essential for the assembly of the SCF^TIR1/AFB2 complex.
(F) Ubiquitination and Glycosylation Crosstalk: Specific F-box proteins (Fbs) of the SCF-type E3 ligases, such as Fbs1 (F-box protein recognizing glycan 1), recognize glycosylated substrates and mediate their ubiquitination and degradation. Red arrows indicate the outcomes of modifications, while black arrows represent enzymatic reactions.
Understanding the types of ubiquitination-centric modification crosstalk provides insight into their regulatory roles in plants. The following sections will elaborate on specific case studies from the literature to further elucidate these concepts.
2.1 Ubiquitination and Phosphorylation: Crucial Factors in Plant Immune Regulation
The innate immune mechanism in plants relies on pattern recognition receptors (PRRs) on the cell membrane to recognize pathogen-associated molecular patterns (PAMPs), thereby triggering pattern-triggered immunity (PTI) responses. During PTI, the generation of reactive oxygen species (ROS) is vital for activating immune responses, a process primarily mediated by members of the nicotinamide adenine dinucleotide phosphate (NADPH) oxidase family, such as RBOHD. Positioned on the membrane, RBOHD possesses six conserved transmembrane helical structures, with its N- and C-termini situated intracellularly.
Upon PAMP recognition by PRRs, rapid influx of Ca2+ triggers conformational changes and phosphorylation of the N-terminus EF-hand motif of RBOHD, ultimately facilitating ROS generation. Despite numerous studies highlighting the pivotal role of intracellular kinases in phosphorylating RBOHD in mediating ROS production, how plants inhibit RBOHD-mediated ROS generation prior to PRR recognition of PAMPs remains a scientifically unresolved question.
In March 2020, the research group led by Gitta Coaker at the University of California, USA, published a research paper titled "Regulation of reactive oxygen species species in plant immunity by phosphorylation and ubiquitination of RBOHD" in the journal Nature Communications. In this paper, through a series of experimental approaches including Co-immunoprecipitation, MBP Pull-down assays, and in vitro phosphorylation assays, the authors confirmed the interaction between PBL13 and the C-terminus of RBOHD, with the C-terminus of PBL13 being crucial for the phosphorylation process of RBOHD.
Furthermore, the authors generated transgenic Arabidopsis thaliana lines with PBL13 knocked out, and through Western blot experiments, observed that knocking out PBL13 increased the accumulation and stability of RBOHD in vivo. Additionally, the study revealed the ubiquitination of RBOHD in vivo. These findings provide important clues for a deeper understanding of the regulatory roles of ubiquitination and phosphorylation in plant immune mechanisms.
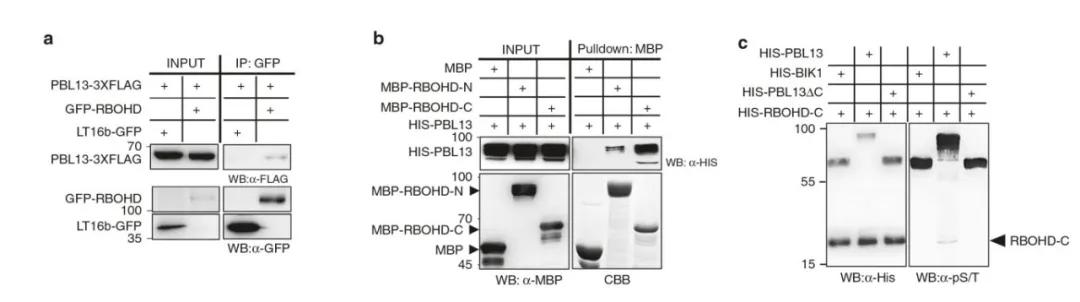 Figure 3 illustrates the binding of PBL13 to the C-terminus of RBOHD, mediating its phosphorylation (Lee et al., 2020). Panel (a) depicts the verification of the interaction between PBL13 and RBOHD through Co-immunoprecipitation. Panel (b) demonstrates the preferential binding of PBL13 to the C-terminus (RBOHD-C) of RBOHD, as visualized by Coomassie Brilliant Blue staining gel. Panel (c) highlights the indispensability of the C-terminus of PBL13 for phosphorylating RBOHD. Previously, BIK1 has been shown to interact with the N-terminus of RBOHD, leading to its phosphorylation (Li et al., 2014).
Figure 3 illustrates the binding of PBL13 to the C-terminus of RBOHD, mediating its phosphorylation (Lee et al., 2020). Panel (a) depicts the verification of the interaction between PBL13 and RBOHD through Co-immunoprecipitation. Panel (b) demonstrates the preferential binding of PBL13 to the C-terminus (RBOHD-C) of RBOHD, as visualized by Coomassie Brilliant Blue staining gel. Panel (c) highlights the indispensability of the C-terminus of PBL13 for phosphorylating RBOHD. Previously, BIK1 has been shown to interact with the N-terminus of RBOHD, leading to its phosphorylation (Li et al., 2014).
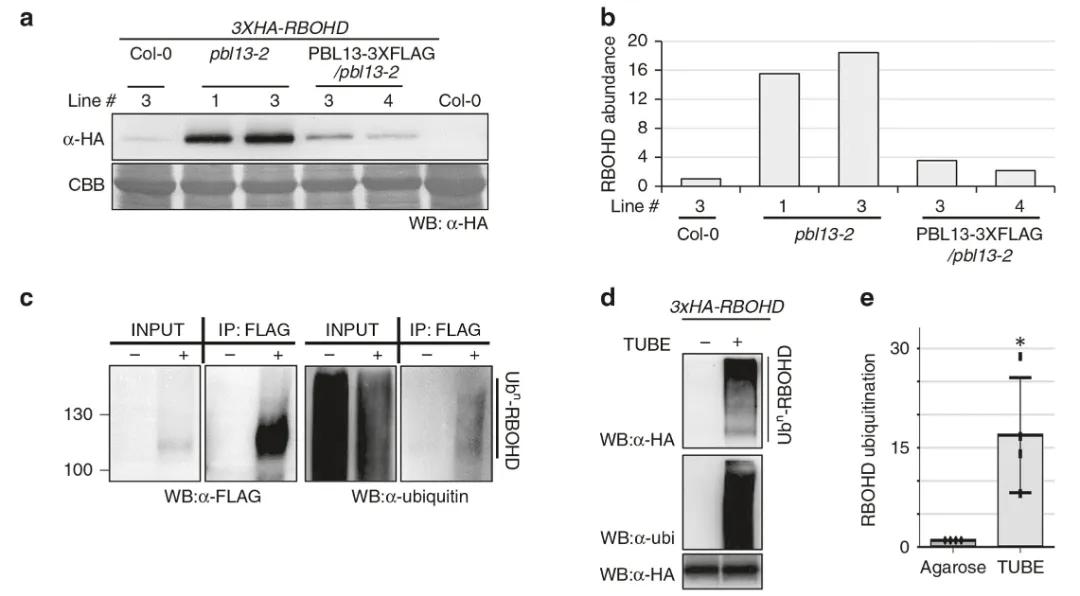 Figure 4 illustrates the inhibition of RBOHD accumulation by PBL13 and the in vivo ubiquitination of RBOHD (Lee et al., 2020). Panel (a) demonstrates reduced levels of RBOHD protein upon PBL13 knockout. Col-0 represents the wild type, plb13-2 denotes the knockout mutant, and PBL13-3×FLAG/plb13-2 signifies the complementation. Panel (b) quantifies the bands from panel (a). Panel (c) depicts the ubiquitination of RBOHD in vivo, with "+" indicating 3×FLAG-RBOHD and "-" representing Col-0. Panel (d) showcases the detection of RBOHD ubiquitination using Tandem Ubiquitin Binding Entities (TUBE), with agarose resin (-) serving as the control. Panel (e) presents the quantification of ubiquitination based on the TUBE method.
Figure 4 illustrates the inhibition of RBOHD accumulation by PBL13 and the in vivo ubiquitination of RBOHD (Lee et al., 2020). Panel (a) demonstrates reduced levels of RBOHD protein upon PBL13 knockout. Col-0 represents the wild type, plb13-2 denotes the knockout mutant, and PBL13-3×FLAG/plb13-2 signifies the complementation. Panel (b) quantifies the bands from panel (a). Panel (c) depicts the ubiquitination of RBOHD in vivo, with "+" indicating 3×FLAG-RBOHD and "-" representing Col-0. Panel (d) showcases the detection of RBOHD ubiquitination using Tandem Ubiquitin Binding Entities (TUBE), with agarose resin (-) serving as the control. Panel (e) presents the quantification of ubiquitination based on the TUBE method.
Subsequently, the authors identified the E3 ubiquitin ligase PIRE, which interacts with the C-terminus of both PBL13 and RBOHD, using protein microarray technology. In vitro ubiquitination assays demonstrated the specific ubiquitination of RBOHD by PIRE, mediating its degradation (Figure 5a, b). Additionally, in vitro and in vivo phosphorylation experiments revealed that phosphorylation of RBOHD enhances its own ubiquitination (Figure 5c-e). The authors evaluated the accumulation of RBOHD in pire gene knockout lines and observed an increase in RBOHD levels in these mutant lines.
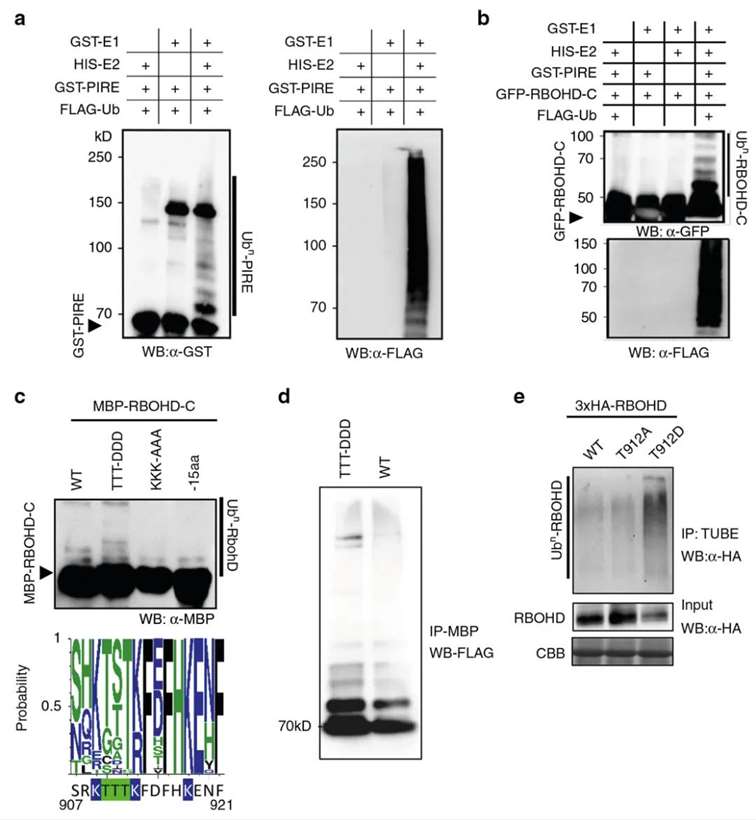 Figure 5 demonstrates the direct ubiquitination of RBOHD by PIRE, leading to the inhibition of RBOHD protein accumulation (Lee et al., 2020). Panel (a) showcases the E3 ubiquitin ligase activity of PIRE. Panel (b) illustrates the direct ubiquitination of RBOHD-C by PIRE. Panel (c) depicts enhanced ubiquitination of RBOHD following in vitro simulation of phosphorylation at the C-terminus. In the figure, "WT" represents the wild type, "TTT-DDD" denotes the phosphomimetic mutant T910DT911DT912D, "KKK-AAA" signifies the lysine mutant K909AK013AK918A, and "-15aa" indicates deletion of 15 amino acids from the C-terminus of RBOHD. Panel (d) confirms the stepwise dependency of ubiquitination through reconstruction of ubiquitination reactions using the phosphomimetic mutant T910DT911DT912D. Panel (e) demonstrates enhanced ubiquitination of RBOHD in vivo with the phosphomimetic RBOHDT912D. Finally, panel (f) presents PIRE's negative regulation of RBOHD protein stability.
Figure 5 demonstrates the direct ubiquitination of RBOHD by PIRE, leading to the inhibition of RBOHD protein accumulation (Lee et al., 2020). Panel (a) showcases the E3 ubiquitin ligase activity of PIRE. Panel (b) illustrates the direct ubiquitination of RBOHD-C by PIRE. Panel (c) depicts enhanced ubiquitination of RBOHD following in vitro simulation of phosphorylation at the C-terminus. In the figure, "WT" represents the wild type, "TTT-DDD" denotes the phosphomimetic mutant T910DT911DT912D, "KKK-AAA" signifies the lysine mutant K909AK013AK918A, and "-15aa" indicates deletion of 15 amino acids from the C-terminus of RBOHD. Panel (d) confirms the stepwise dependency of ubiquitination through reconstruction of ubiquitination reactions using the phosphomimetic mutant T910DT911DT912D. Panel (e) demonstrates enhanced ubiquitination of RBOHD in vivo with the phosphomimetic RBOHDT912D. Finally, panel (f) presents PIRE's negative regulation of RBOHD protein stability.
To investigate the significance of PIRE in plant innate immunity, the authors initially examined whether PIRE undergoes post-translational modifications before and after flg22 treatment. The results indicated dynamic phosphorylation modifications of PIRE following treatment with flg22 (Figure 6). Subsequently, the authors generated two gene knockout transgenic lines, PIRE-1 and PIRE-2, and assessed the levels of RBOHD and reactive oxygen species (ROS) in wild-type plants and transgenic lines before and after flg22 treatment. It was observed that the levels of both RBOHD and ROS increased in the transgenic lines following flg22 treatment compared to the wild-type plants.
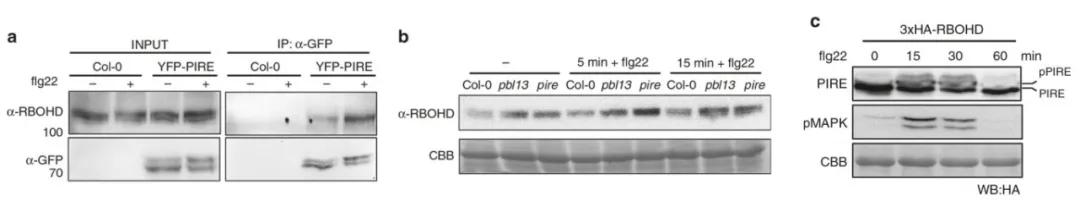 Figure 6 illustrates the interaction between PIRE and RBOHD, with immune activation resulting in dynamic phosphorylation of PIRE (Lee et al., 2020). Panel (a) demonstrates the binding of PIRE to RBOHD following immune activation. "+" denotes treatment with flg22, while "-" indicates no flg22 treatment. Panel (b) shows enhanced expression of RBOHD following flg22 treatment in pbl13 and pire knockout lines. Panel (c) depicts changes in the phosphorylation status of PIRE upon perception of PAMPs.
Figure 6 illustrates the interaction between PIRE and RBOHD, with immune activation resulting in dynamic phosphorylation of PIRE (Lee et al., 2020). Panel (a) demonstrates the binding of PIRE to RBOHD following immune activation. "+" denotes treatment with flg22, while "-" indicates no flg22 treatment. Panel (b) shows enhanced expression of RBOHD following flg22 treatment in pbl13 and pire knockout lines. Panel (c) depicts changes in the phosphorylation status of PIRE upon perception of PAMPs.
In summary, prior to pathogen perception, PIRE constitutively binds to RBOHD, inhibiting the accumulation of RBOHD protein through phosphorylation and ubiquitination. However, upon pathogen perception (flg22), RBOHD becomes activated, leading to dynamic phosphorylation of PIRE, thereby regulating the generation of RBOHD-dependent reactive oxygen species (ROS) (Figure 7).
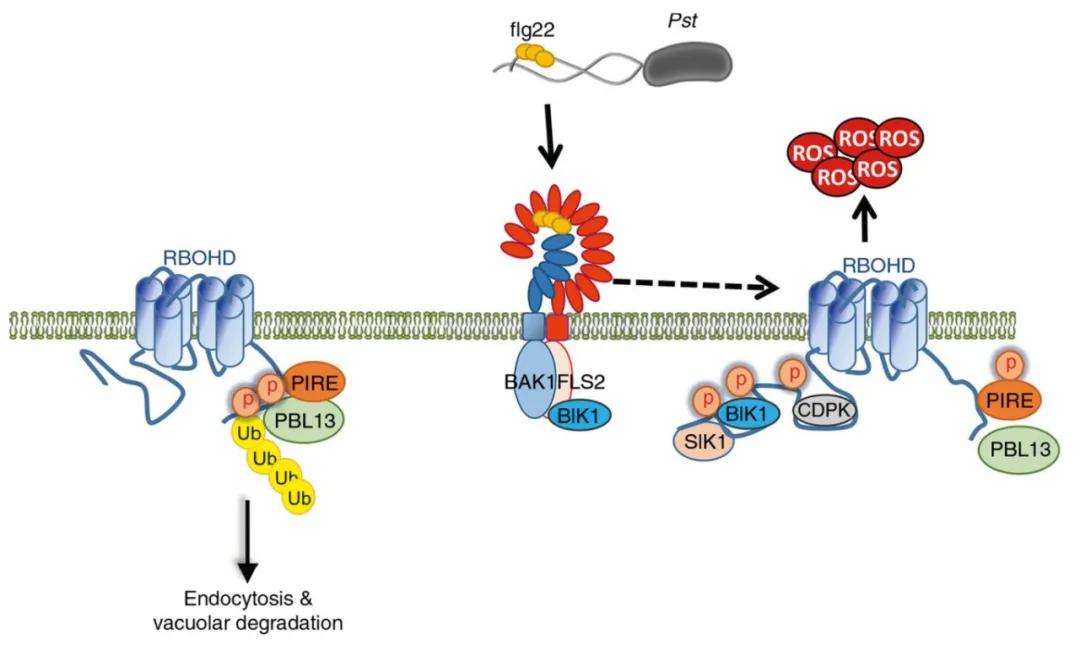 Figure 7 Model diagram of PBL13 and PIRE regulating RBOHD (Lee et al., 2020).
Figure 7 Model diagram of PBL13 and PIRE regulating RBOHD (Lee et al., 2020).
2.2 Poly(ADP-ribosyl)ation and K63-Linked Ubiquitination: Regulating Plant Immunity
Poly(ADP-ribosyl)ation (PARylation) is a reversible post-translational modification (PTM) catalyzed by ADP-ribosyl transferases (PARPs) and ADP-ribosyl hydrolases (PARGs). PARPs transfer ADP-ribose onto specific amino acid residues of substrate proteins, forming poly(ADP-ribose) chains, while PARGs hydrolyze ADP-ribose from substrate proteins. Although PARylation and ubiquitination can independently mediate various biochemical processes, their crosstalk in regulating plant immunity remains unclear. In August 2021, the research group led by Junqi Song at Texas A&M University published a study titled "Coordinated Regulation of Plant Immunity by Poly(ADP-ribosyl)ation and K63-Linked Ubiquitination" in the journal Molecular Plant. This study demonstrates that Arabidopsis UBC13A and UBC13B are the main drivers of K63-linked polyubiquitin chain-mediated ubiquitination. They directly interact with PARPs and PARGs, collectively regulating plant immune responses.
The authors initially identified a protein, UBC13, interacting with PARPs and PARGs through yeast two-hybrid screening. UBC13 is a ubiquitin-conjugating enzyme catalyzing the synthesis of K63-linked polyubiquitin chains (Pastushok et al., 2005). The Arabidopsis genome encodes two UBC13 genes, namely UBC13A and UBC13B. Subsequent in vitro and in vivo PARylation experiments revealed that UBC13B undergoes PARP2-mediated PARylation modification, with increased modification levels upon flg22 treatment (Figure 8A, B). Through proteomics and in vitro PARylation experiments, four glutamic acid residues (E31/E32/E62/E63) in UBC13B were identified as PARylation sites (Figure 8C-E). Mutating these residues simultaneously to alanine (A) yielded the UBC13B4E mutant, which, in co-immunoprecipitation experiments, exhibited reduced interaction strength with PARP2, indicating that PARylation promotes the interaction between UBC13B and PARP2 (Figure 8F). Observations of Arabidopsis phenotypes after introducing UBC13B4E into ubc13a/ubc13b double mutants revealed that while the plant size resembled the wild type, it remained susceptible to disease, suggesting that UBC13B PARylation participates in regulating plant immunity (Figure 8G, H).
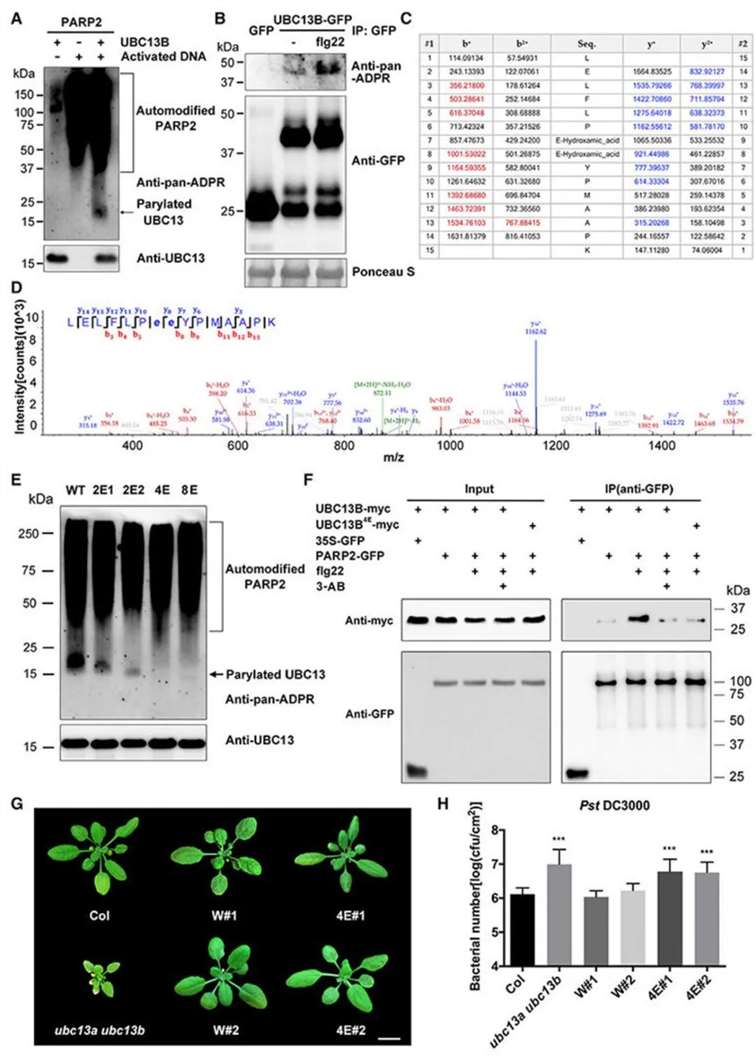 Figure 8: Involvement of UBC13B PARylation in Regulating Plant Immunity (Yao et al., 2021). (A) In vitro PARylation assay; (B) Elevated PARylation level of UBC13B upon flg22 treatment; (C) Mass-to-charge ratio list of b+ and y+ ions for peptide (LELFLPEEYPMAAPK), where red and blue mass values correspond to identified peaks of b+ and y+ ions in the spectrum; (D) MS/MS spectrum of doubly charged peptides with PARylated Glu residues. Bold and lowercase "e" denote modified Glu residues; (E) Significant reduction in UBC13B PARylation by glutamic acid mutations in vitro. UBC13B2E1 (E31/E32), UBC13B2E2 (E62/E63), UBC13B4E (E31/E32/E62/E63), and UBC13B8E (all eight Glu residues replaced with alanine at the N-terminus of UBC13B); (F) Reduced interaction between UBC13B4E and PARP2; (G) Phenotypic analysis. Col: wild type, ubc13a/ubc13b: double mutant, W#1: UBC13B introduced into Col, W#2: UBC13B introduced into ubc13a/ubc13b double mutant, 4E#1: UBC13B4E introduced into Col, 4E#2: UBC13B4E introduced into ubc13a/ubc13b double mutant; (H) Content of Pseudomonas syringae pv. tomato (Pst DC3000) in UBC13B4E mutant plants.
Figure 8: Involvement of UBC13B PARylation in Regulating Plant Immunity (Yao et al., 2021). (A) In vitro PARylation assay; (B) Elevated PARylation level of UBC13B upon flg22 treatment; (C) Mass-to-charge ratio list of b+ and y+ ions for peptide (LELFLPEEYPMAAPK), where red and blue mass values correspond to identified peaks of b+ and y+ ions in the spectrum; (D) MS/MS spectrum of doubly charged peptides with PARylated Glu residues. Bold and lowercase "e" denote modified Glu residues; (E) Significant reduction in UBC13B PARylation by glutamic acid mutations in vitro. UBC13B2E1 (E31/E32), UBC13B2E2 (E62/E63), UBC13B4E (E31/E32/E62/E63), and UBC13B8E (all eight Glu residues replaced with alanine at the N-terminus of UBC13B); (F) Reduced interaction between UBC13B4E and PARP2; (G) Phenotypic analysis. Col: wild type, ubc13a/ubc13b: double mutant, W#1: UBC13B introduced into Col, W#2: UBC13B introduced into ubc13a/ubc13b double mutant, 4E#1: UBC13B4E introduced into Col, 4E#2: UBC13B4E introduced into ubc13a/ubc13b double mutant; (H) Content of Pseudomonas syringae pv. tomato (Pst DC3000) in UBC13B4E mutant plants.
The PR1 gene serves as a widely recognized marker of defense responses in plants (Tsuda et al., 2008). Consequently, the authors assessed the expression levels of PR1 in parp and ubc13a/ubc13b mutants, revealing an increase in PR1 content. However, extracellular secretion of PR1 showed a decreasing trend compared to the wild type (Figure 9A). Studies have linked numerous proteins involved in the secretory pathway, such as PDI2 and SYP, to K63-mediated ubiquitination (Romero-Barrios et al., 2020). Hence, the authors speculated that PDI PARylation and K63-mediated ubiquitination might influence the secretion of PR1 protein. To validate this hypothesis, the authors conducted Y2H experiments confirming interactions between PDI and PARP2 as well as PARG1 (Figure 9B). Furthermore, Western blot analysis demonstrated both PARylation and K63-mediated ubiquitination of PDI, with PARylation of PDI shown to enhance its own ubiquitination level (Figure 9C).
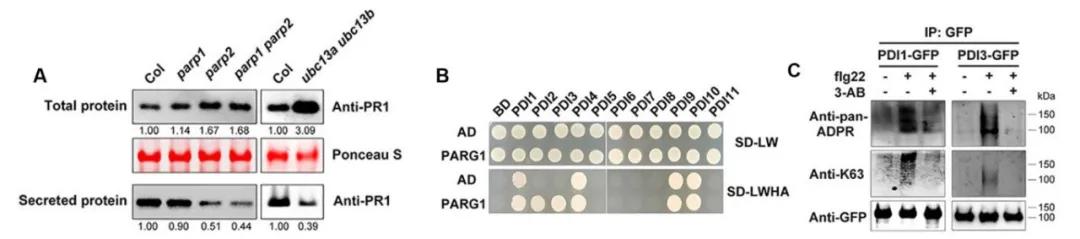 Figure 9 illustrates the impact of PDI PARylation and K63-mediated ubiquitination on the secretion process of PR1 protein (Yao et al., 2021). Panel (A) displays the total PR1 protein content and secreted PR1 protein levels in parp and ubc13 mutants. Panel (B) demonstrates the interaction between PDI2/3 and PARG validated through yeast two-hybrid (Y2H) assay. Panel (C) exhibits flg22-induced PARylation of PDI and enhanced K63-linked ubiquitination of intracellular PDI.
Figure 9 illustrates the impact of PDI PARylation and K63-mediated ubiquitination on the secretion process of PR1 protein (Yao et al., 2021). Panel (A) displays the total PR1 protein content and secreted PR1 protein levels in parp and ubc13 mutants. Panel (B) demonstrates the interaction between PDI2/3 and PARG validated through yeast two-hybrid (Y2H) assay. Panel (C) exhibits flg22-induced PARylation of PDI and enhanced K63-linked ubiquitination of intracellular PDI.
The aforementioned findings elucidate that, in the PAMP response, PARP proteins recruit UBC13 and PDI through interaction and PARylation, thereby facilitating UBC13-mediated K63 ubiquitination of PDI, leading to PDI activation. Subsequently, PDI collaborates with other secretory proteins to fulfill its distinct function, ensuring proper protein folding and secretion into the extracellular space (Figure 10), ultimately contributing to the regulation of plant immune responses.
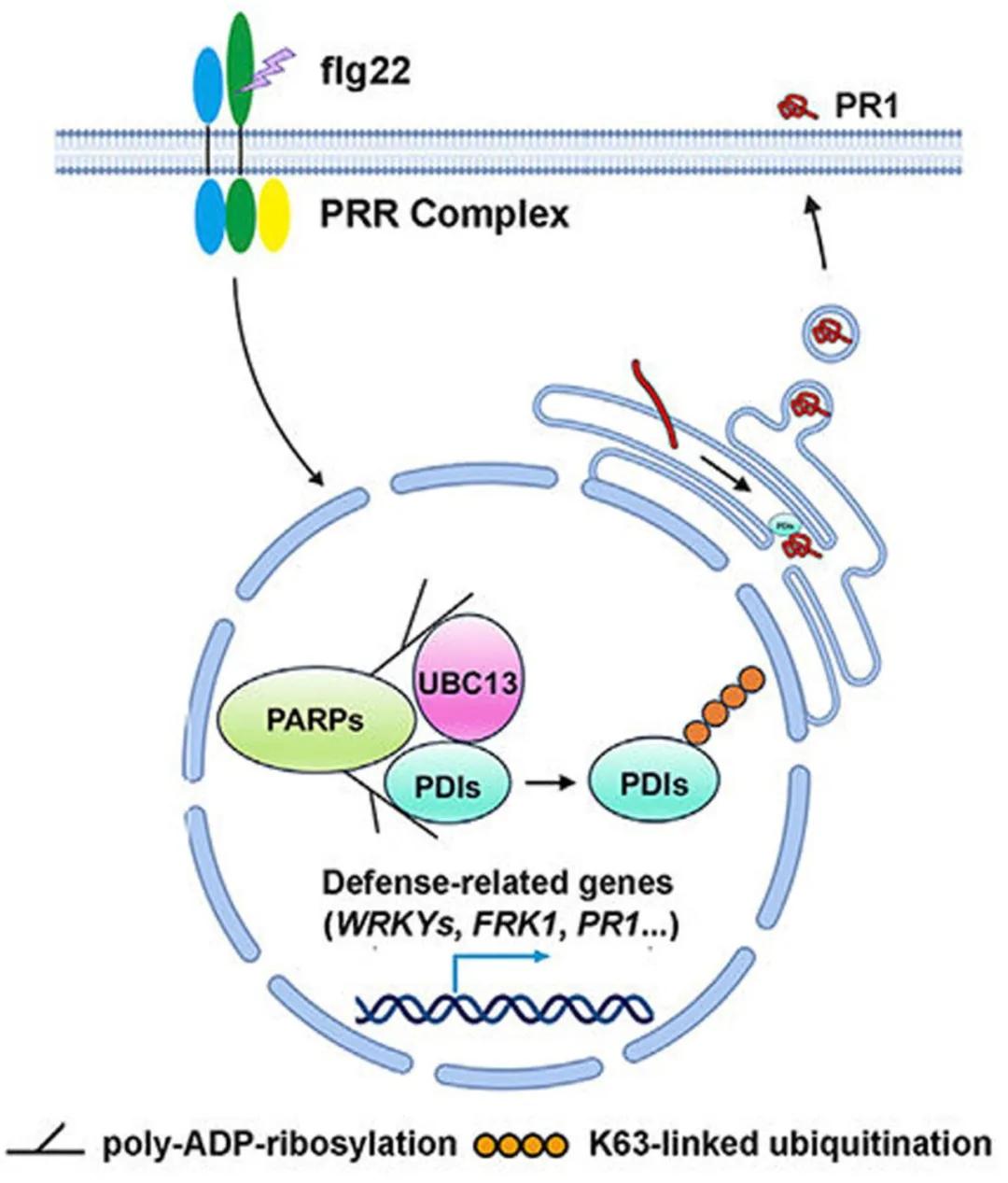 Figure 10 A model in which PARylation and K63-mediated ubiquitination coordinately regulate the secretory pathway (Yao et al., 2021).
Figure 10 A model in which PARylation and K63-mediated ubiquitination coordinately regulate the secretory pathway (Yao et al., 2021).
2.3 Ubiquitination and Acetylation - Regulating Changes in Plant Protein Stability
Dynamic changes in the proteome are crucial mechanisms for plants to adapt to environmental variations, with alterations in protein stability playing a pivotal role in plant growth and development. The ubiquitin-proteasome system (UPS) is involved in the degradation of the majority of proteins within plants; however, how PTMs regulate protein stability remains elusive. In February 2022, Markus Wirtz's group at Heidelberg University published a research article titled "Cotranslational N-degron masking by acetylation promotes proteome stability in plants" in the journal Nature Communications. In this study, the authors demonstrated that damage to N-terminal acetylation (NTA) dependent on NatA leads to global instability in the Arabidopsis proteome and identified a novel degradation signal marking most non-acetylated cytoplasmic proteins for degradation via the ubiquitin system.
Loss of NatA results in embryonic lethality in plants (Linster et al., 2015). Hence, by employing amiRNA-mediated interference with the expression of NAA10 and NAA15 subunits of NatA, the authors observed a fourfold increase in protein degradation rate and enhanced ubiquitin-proteasome system (UPS) activity upon interference (Figure 11). Subsequently, the authors evaluated the total protein content in damaged plant leaves due to NatA disruption and found no significant impact on total protein levels (Figure 12a). Proteomic analysis revealed that only 92 proteins exhibited significant changes in NatA-damaged plants, with 83% identified as NatA substrates (Figure 12b). Two proteins identified as NatA substrates (significantly downregulated ARKGR1 and non-significantly upregulated ASROAS-TLA) were selected for protein stability assays, demonstrating degradation of both proteins in NatA-damaged plants (Figure 12c, d). However, upon treatment with the proteasome inhibitor MG132, ASROAS-TLA rapidly accumulated in NatA-damaged plants, indicating that the unaffected stability of ASROAS-TLA was due to enhanced translation, while the stability of non-NatA substrates MPPSAT5 and MRETUBB4 remained unaffected (Figure 12e).
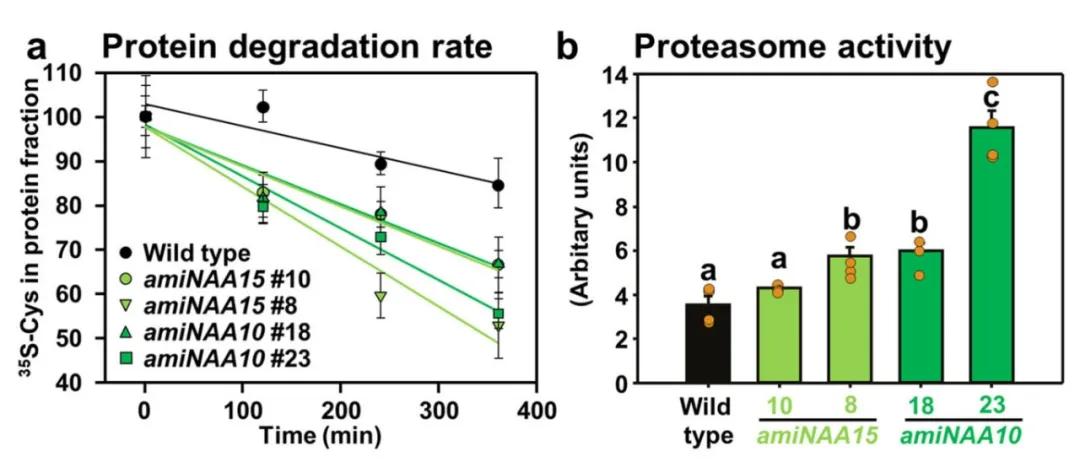 Figure 11 Damage to NatA will increase ubiquitin-proteasome activity and lead to protein degradation (Linster et al., 2022).
Figure 11 Damage to NatA will increase ubiquitin-proteasome activity and lead to protein degradation (Linster et al., 2022).
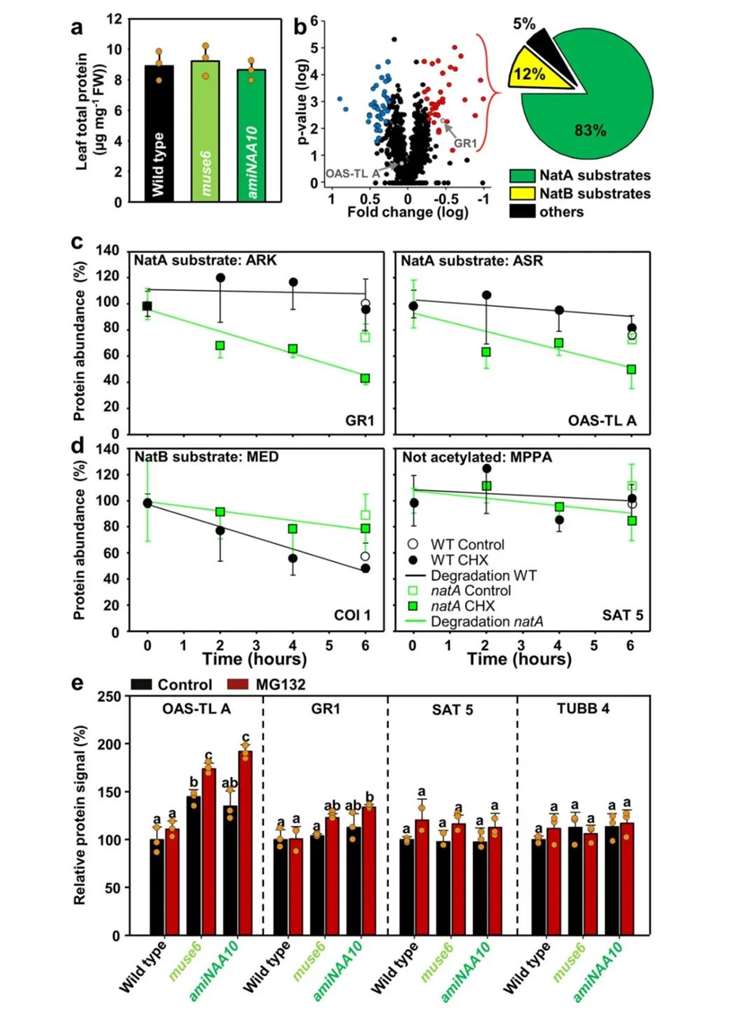 Figure 12 demonstrates that NatA damage does not affect total protein concentration but significantly disrupts the stability of NatA substrates (Linster et al., 2022). Panel (a) depicts the concentration of total proteins in leaves of both wild-type and NatA-damaged plants. Panel (b) presents a volcano plot illustrating the differential expression of proteins in wild-type leaves compared to amiNAA10-expressing leaves. Red denotes reduction, while blue signifies accumulation, with the pie chart indicating the classification of NatA substrates among the significantly reduced proteins. Panels (c, d) depict the degradation analysis of NatA substrates (GR1, OAS-TLA) and N-terminal proteins not acetylated by NatA (COI1, SAT5). CHX, cycloheximide, a translation inhibitor, is used. Panel (e) illustrates the relative levels of OAS-TLA, GR1, SAT5, and TUBB4 proteins in leaves of wild-type and NatA-damaged plants. MG132 denotes the proteasome inhibitor.
Figure 12 demonstrates that NatA damage does not affect total protein concentration but significantly disrupts the stability of NatA substrates (Linster et al., 2022). Panel (a) depicts the concentration of total proteins in leaves of both wild-type and NatA-damaged plants. Panel (b) presents a volcano plot illustrating the differential expression of proteins in wild-type leaves compared to amiNAA10-expressing leaves. Red denotes reduction, while blue signifies accumulation, with the pie chart indicating the classification of NatA substrates among the significantly reduced proteins. Panels (c, d) depict the degradation analysis of NatA substrates (GR1, OAS-TLA) and N-terminal proteins not acetylated by NatA (COI1, SAT5). CHX, cycloheximide, a translation inhibitor, is used. Panel (e) illustrates the relative levels of OAS-TLA, GR1, SAT5, and TUBB4 proteins in leaves of wild-type and NatA-damaged plants. MG132 denotes the proteasome inhibitor.
Based on the foregoing findings, the authors proposed a hypothesis that the significant upregulation of translation in NatA-damaged plants could maintain proteome stability and conducted relevant validation experiments. The results indicated a fourfold increase in protein translation in NatA-deficient plants, attributed to selective translation enhancement of various proteins (Figure 13a, b). The Target of Rapamycin (TOR) kinase is a pivotal regulator of ribosome biogenesis, modulating ribosome quantity and the translation efficiency of stress-related genes via phosphorylation of S6 kinase and translation initiation factor eIF3h (Dong et al., 2017). Damage to NatA led to increased TOR activity, resulting in significant rRNA accumulation (Figure 13c, d). Subsequently, the authors employed dual orthogonal noncanonical amino acid tagging experiments (Glenn et al., 2017) to demonstrate higher protein translation rates in NatA-damaged plants, with 72% of the proteins being NatA substrates (Figure 13e-g). This suggests that the selective instability caused by N-terminal acetylation impairment in NatA substrates is counteracted by enhanced translation, maintaining their steady-state levels in the mutant lines.
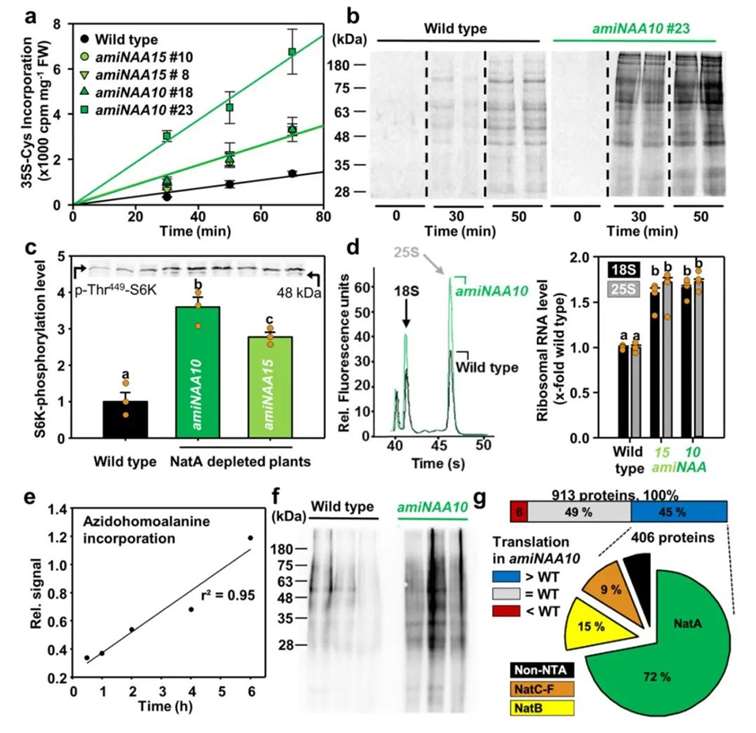 Figure 13 illustrates that NatA-damaged plants exhibit a higher translation rate of NatA substrates, promoted by TOR-induced ribosome biogenesis (Linster et al., 2022). Panel (a) depicts the incorporation of isotopically labeled sulfur amino acids into leaf proteins of both wild-type and NatA-damaged plants. Panel (b) shows SDS-PAGE separation of proteins from wild-type and amiNAA10 leaves at specified time points post-incorporation of isotopically labeled sulfur amino acids. Panel (c) quantifies the phosphorylation of TOR target S6KT449 in leaves of wild-type and NatA-damaged plants using phospho-specific antibodies. Panels (d) quantify 18S and 25S rRNA levels in leaves of wild-type and NatA-damaged plants. Panel (e) validates the content of linear azidohomoalanine in proteins from wild-type leaves. Panel (f) compares the incorporation of azidohomoalanine into leaf proteins of wild-type and NatA-damaged plants three hours post-biotin labeling mediated by azidohomoalanine. Panel (g) presents proteomic analysis of newly synthesized proteins in leaves of wild-type and amiNAA10 plants, with the pie chart depicting the classification of NatA substrates in the fraction of proteins translated more efficiently in amiNAA10 plants.
Figure 13 illustrates that NatA-damaged plants exhibit a higher translation rate of NatA substrates, promoted by TOR-induced ribosome biogenesis (Linster et al., 2022). Panel (a) depicts the incorporation of isotopically labeled sulfur amino acids into leaf proteins of both wild-type and NatA-damaged plants. Panel (b) shows SDS-PAGE separation of proteins from wild-type and amiNAA10 leaves at specified time points post-incorporation of isotopically labeled sulfur amino acids. Panel (c) quantifies the phosphorylation of TOR target S6KT449 in leaves of wild-type and NatA-damaged plants using phospho-specific antibodies. Panels (d) quantify 18S and 25S rRNA levels in leaves of wild-type and NatA-damaged plants. Panel (e) validates the content of linear azidohomoalanine in proteins from wild-type leaves. Panel (f) compares the incorporation of azidohomoalanine into leaf proteins of wild-type and NatA-damaged plants three hours post-biotin labeling mediated by azidohomoalanine. Panel (g) presents proteomic analysis of newly synthesized proteins in leaves of wild-type and amiNAA10 plants, with the pie chart depicting the classification of NatA substrates in the fraction of proteins translated more efficiently in amiNAA10 plants.
Post-translational modifications of proteins have long been a focal point in life sciences research, garnering significant attention. From common phosphorylation and ubiquitination modifications to less prevalent palmitoylation modifications, numerous studies have aimed to delve into the specific roles of individual modifications in biological activities. This paper extensively enumerates reported instances of crosstalk between ubiquitination and other modifications in plants. It systematically elucidates the synergistic roles of ubiquitination with phosphorylation and poly(ADP-ribosyl)ation in jointly regulating plant immune responses, as well as the molecular mechanisms underlying the crosstalk between ubiquitination and acetylation in modulating changes in protein stability within organisms.
Summary
Although crosstalk between modifications may sound complex, the research methodologies are, in fact, quite similar to those used in studying individual modifications. Researchers can identify interacting proteins as entry points, subsequently verifying whether the target protein undergoes corresponding modifications, thereby elucidating relevant biological functions. The modifications crosstalk and their applications listed in this paper represent only the tip of the iceberg. In reality, the types and functions of modification crosstalk are far more extensive, awaiting further exploration and discovery by the scientific community.
References
- Venne A S, Kollipara L, Zahedi R P. The next level of complexity: crosstalk of posttranslational modifications. Proteomics, 2014, 14(4-5): 513-524.
- Hunter T. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Molecular cell, 2007, 28(5): 730-738.
- Zhou B, Zeng L. Conventional and unconventional ubiquitination in plant immunity. Molecular plant pathology, 2017, 18(9): 1313-1330.
- Zhao Y, Brickner J R, Majid M C, et al. Crosstalk between ubiquitin and other post-translational modifications on chromatin during double-strand break repair. Trends in cell biology, 2014, 24(7): 426-434.
- Zhang Y, Zeng L. Crosstalk between ubiquitination and other post-translational protein modifications in plant immunity. Plant Communications, 2020, 1(4).
- Averyanov A. Oxidative burst and plant disease resistance. Front Biosci (Elite Ed), 2009, 1: 142-152.
- Dubiella U, Seybold H, Durian G, et al. Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proceedings of the National Academy of Sciences, 2013, 110(21): 8744-8749.
- Lee D H, Lal N K, Lin Z J D, et al. Regulation of reactive oxygen species during plant immunity through phosphorylation and ubiquitination of RBOHD. Nature Communications, 2020, 11(1): 1838.
- Li L, Li M, Yu L, et al. The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell host & microbe, 2014, 15(3): 329-338.
- Yao D, Arguez M A, He P, et al. Coordinated regulation of plant immunity by poly (ADP-ribosyl) ation and K63-linked ubiquitination. Molecular Plant, 2021, 14(12): 2088-2103.
- Tsuda K, Sato M, Glazebrook J, et al. Interplay between MAMP‐triggered and SA‐mediated defense responses. The Plant Journal, 2008, 53(5): 763-775.
- Romero-Barrios N, Monachello D, Dolde U, et al. Advanced cataloging of lysine-63 polyubiquitin networks by genomic, interactome, and sensor-based proteomic analyses. The Plant Cell, 2020, 32(1): 123-138.
- Linster E, Forero Ruiz F L, Miklankova P, et al. Cotranslational N-degron masking by acetylation promotes proteome stability in plants. Nature Communications, 2022, 13(1): 810.
- Linster E, Stephan I, Bienvenut W V, et al. Downregulation of N-terminal acetylation triggers ABA-mediated drought responses in Arabidopsis. Nature Communications, 2015, 6(1): 7640.
- Pastushok L, Moraes T F, Ellison M J, et al. A single Mms2 "key" residue insertion into a Ubc13 pocket determines the interface specificity of a human Lys63 ubiquitin conjugation complex. Journal of Biological Chemistry, 2005, 280(18): 17891-17900.
- Glenn W S , Stone S E , Ho S H ,et al.Bioorthogonal Noncanonical Amino Acid Tagging (BONCAT) Enables Time-Resolved Analysis of Protein Synthesis in Native Plant Tissue.PLANT PHYSIOLOGY, 2017, 173(3):1543-1553.
- Dong Y , Silbermann M , Speiser A ,et al.Sulfur availability regulates plant growth via glucose-TOR signaling.Nature Communications, 2017, 8(1):1174
Our products and services are for research use only.