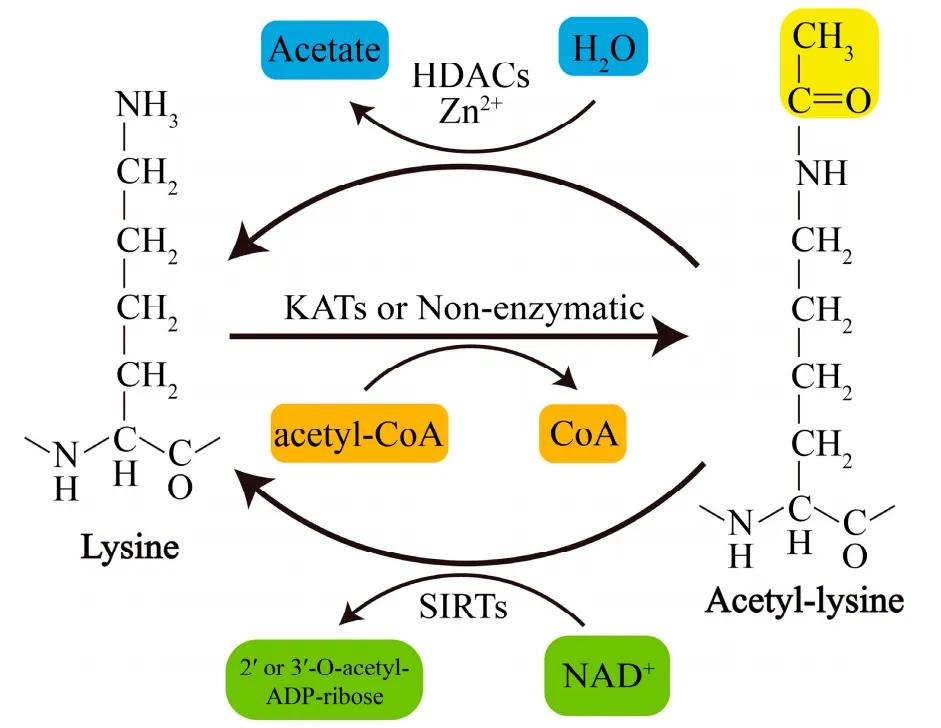
Acetylation serves as a significant modality of post-translational modification within cells. Proteins, including histones, are often subjected to this molecular process.
The Dynamical Acetylation Modification of Histones
The dynamic acetylation modification of histones is well-examined over recent years. First discovered over 40 years ago, reversible acetylation occurs on the lysines present at the N-terminal of core histones. The acetylation of histones is catalyzed by histone acetyltransferases (HATs), and deacetylation is catalyzed by histone deacetylases (HDACs or HDs).
The heavily lysine-enriched tail of the core histones carries a positive charge under physiological conditions, enabling interactions with negatively charged DNA or adjacent nucleosomes. This interaction leads to the compaction of nucleosomal structure and highly folded chromatin. Acetylation weakens the interaction between histones and DNA, leading to a looser chromatin conformation, which aids in the approach of transcription regulators and thus promotes gene transcription. In contrast, deacetylation inhibits gene transcription.
It is postulated that acetylation within the N-terminal domain of histones leads to local chromatin unwinding - a necessitous but not sufficient condition for DNA repackaging. Several different HATs and HDs have now been identified, with CBP/p300 possibly being the most significant, able to interact with multiple transcription-modulating factors. The half-life of histone acetylation and deacetylation is merely a few minutes. The role of these persistent, rapid competitive reactions remains unclear. It is not yet known whether this change itself is related to chromatin transcription. Some studies indicate that this variation might be directly linked to gene transcription.
Select Service
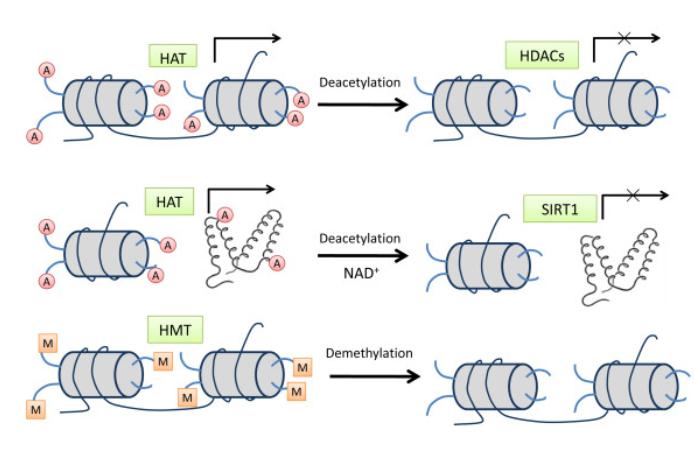 Histone modification pathways. (Yuanyuan Rose Li et al,. 2011)
Histone modification pathways. (Yuanyuan Rose Li et al,. 2011)
Elevated acetylation in normally suppressed regions, or deacetylation in ordinarily transcriptionally active locales, can result in various dysfunctions, giving rise to developmental and proliferative diseases such as leukemia, skin cancer, fragile X syndrome, and thumb malformation syndrome. Harel-Bellan thoroughly investigated diseases arising from abnormalities in acetylation and deacetylation processes, thereby this will not be further elaborated upon here. In vivo, alterations in enhancer activity may regulate the binding capacity of recombinases to chromosomal recombination signal sequences, thereby controlling V(D)J recombination, although the molecular mechanism underlying this change remains unclear. McMurry et al. examined the influence of histone H3 acetylation on V(D)J recombinase reporter genes and T cell receptor α/δ loci. Their empirical findings confirm the long-term regulatory role of enhancer activities in the context of H3 acetylation, and that H3 acetylation levels are closely associated with V(D)J recombination. Therefore, enhanced H3 acetylation may serve as a molecular mechanism linking enhancer activity with the capacity for V(D)J recombination.
Polyglutamine diseases represent a class of neurodegenerative genetic disorders caused by expansions of CAG repeat segments within pathogenic genes. Studies have illuminated that in these expansion-induced disorders, an imbalance in protein acetylation and deacetylation represents a critical process. By leveraging genetic or pharmacological approaches to diminish the relative presentation of enzymatic clusters, such as histone deacetylases (HDACs), equilibrium can be partly restored. As our understanding of HDACs deepens, it becomes feasible to develop inhibitors targeted at specific HDAC family proteins, and these compounds may potentially have therapeutic efficacy in the management of these diseases.
Other Acetylation
In research on the acetylation of microtubule proteins, Hubbert et al. discovered that histone deacetylase 6 (HDAC6) can reverse the post-translational modification of acetylated microtubule proteins. Evidence suggests that a reduction in the acetylation level of microtubule proteins enhances cell motility while simultaneously decreasing the stability of microtubules. However, research conducted by Palazzo and colleagues indicated no direct correlation between microtubule stability and the degree of microtubule protein acetylation. Changes in cell motility are therefore likely driven by variations in the acetylation levels of microtubule proteins.
In 2004, Nguyen and colleagues discovered an association between acetylation of the retinoblastoma protein (pRb) and cellular differentiation processes, including the formation of skeletal muscles, proposing that acetylation could regulate the specific differentiation functionality of pRb. The retinoblastoma protein is implicated in the induction of cell growth arrest and differentiation.
The auxiliary activation factor, p300, is capable of acetylating the retinoblastoma protein, albeit the biological impact of this acetylation remains unclear at this stage. The p300-Associated Factor (P/CAF) possesses an acetyltransferase domain that directly interacts with the retinoblastoma protein and controls its acetylation level in vivo. Interestingly, P/CAFs from different cells can interact with each other as well.
Experimental findings suggest that acetylation does not affect the growth arrest induced by the retinoblastoma protein nor does it affect the transcriptional inhibitory activity of E2F. Conversely, terminal cell cycle arrest and the expression of myogenesis genes mediated by the retinoblastoma protein seem to necessitate acetylation.
References
- Hansen J C, Tse C, Wolffe A P. Structure and function of the core histone N-termini: more than meets the eye. Biochemistry, 1998, 37: 17637~17641
- Walia H, Chen H Y, Sun J M, et al. Histone acetylation is required to maintain the unfolded nucleosome structure associated with transcribing DNA. J Biol Chem, 1998, 17: 2865~2876
- Waterborg J H. Dynamics of histone acetylation in vivo. A function for acetylation turnover? Biochemistry and Cell Biologybiochimie et Biologie Cellulaire, 2002, 80(3): 363~378
- Bodai L, Pallos J, Thompson L M, et al. Altered protein acetylation in polyglutamine diseases. Current Medicinal Chemistry, 2003, 10(23): 2577~2587
- McMurry M T, Krangel M S. A role for histone acetylation in the developmental regulation of V(D)J recombination. Science, 2000, 287: 495~498
- Hubbert C, Amaris G, Shao R, et al. HDAC6 is a microtubuleassociated deacetylase. Nature, 2002, 417(6887): 455~458[DOI] 63 Palazzo A, Ackerman B, Gundersen G G. Tubulin acetylation and cell motility. Nature, 2003, 421: 230
- Nguyen D X, Baglia L A, Huang S M, et al. Acetylation regulates the differentiation-specific functions of the retinoblastoma protein. The EMBO Journal, 2004, 23(7): 1609~1618
Our products and services are for research use only.