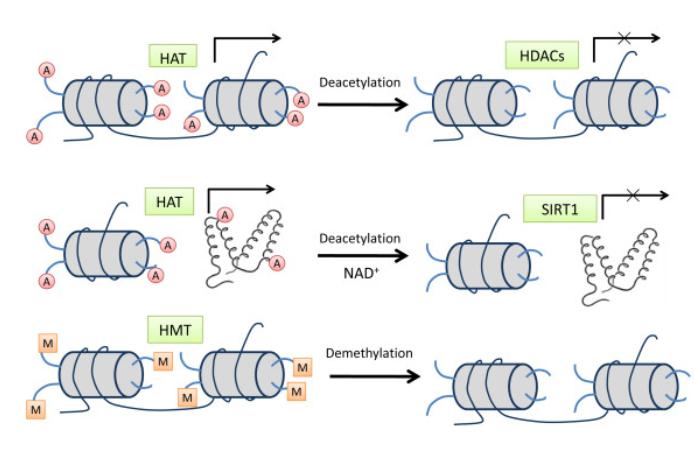
Protein acetylation, as a prevalent post-translational modification found in both eukaryotic and prokaryotic organisms, is evidently ubiquitous and significant. The acetylation process entails the addition of acetyl groups to either the N-terminus of proteins or lysine residues, catalyzed by acetyltransferases or non-biological catalysts. This mechanism plays a crucial role in regulating protein function.
Acetylation modification at the N-terminus of proteins primarily occurs in eukaryotes and is catalyzed by N-terminal acetyltransferases (NATs). It is noteworthy that enzymes capable of executing N-terminal deacetylation have not been discovered to date, hence, N-terminal acetylation is generally deemed irreversible. In contrast, acetylation modification on lysine residues is more widespread, occurring not only in eukaryotes but also extensively distributed in prokaryotes. This process is predominantly catalyzed by lysine acetyltransferases (KATs) and lysine deacetylases (KDACs), forming a reversible acetylation modification cycle.
This article systematically elucidates pertinent knowledge regarding acetylation as a protein post-translational modification. Initially, it introduces the fundamental concepts and background of acetylation modification. Subsequently, it meticulously analyzes three commonly employed methods for protein acetylation identification. Finally, it focuses on the significant role of acetylation modification in plant biological functions, aiming to provide valuable reference and insights for research in related fields.
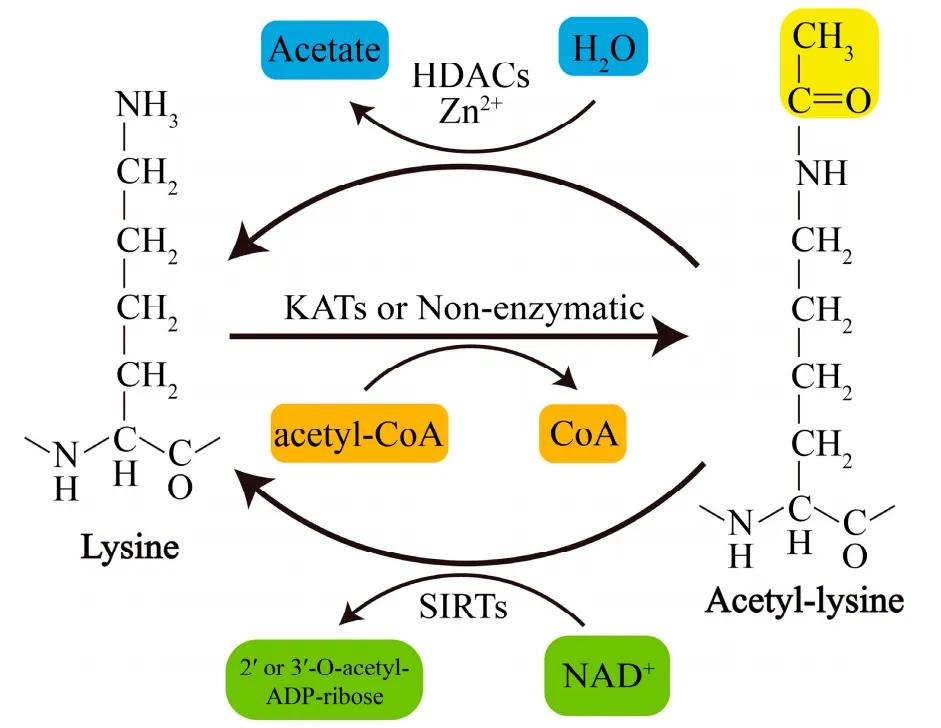 Figure 1 Schematic diagram of the reversible reaction of lysine acetylation (Wang et al., 2020).
Figure 1 Schematic diagram of the reversible reaction of lysine acetylation (Wang et al., 2020).
N-terminal acetyltransferases
N-terminal acetyltransferases (NATs) belong to the GNAT (Gcn5-related N-acetyltransferase) protein family. Based on substrate specificity and subunit composition, they can be categorized into multiple subtypes, including NatA, NatB, NatC, NatD, NatE, NatF, and NatG. Among these, NatA, NatB, and NatC primarily target the N-terminus of proteins and typically comprise a catalytic subunit along with one or two auxiliary subunits.
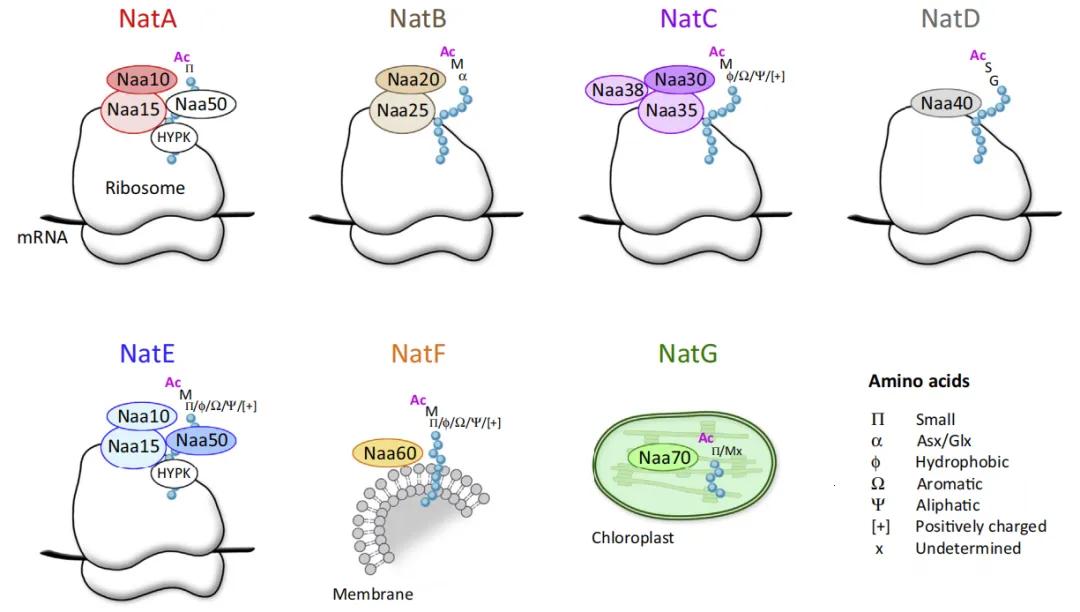 Figure 2 Different types of NATs in eukaryotes (Aksnes et al., 2016).
Figure 2 Different types of NATs in eukaryotes (Aksnes et al., 2016).
Lysine acetyltransferases
KATs encompass three protein families: the GNAT family, comprising KAT2A and KAT2B; the MYST family, including MOZ, Ybf2/Sas3, Sas2, and Tip60; and the CBP/p300 family. KAT2A and KAT2B, highly homologous acetyltransferases within the GNAT family, exhibit mutual exclusion within chromatin modification complexes (Nagy et al., 2010; Foumier et al., 2016). Tip60, a extensively studied KAT in the MYST family, plays pivotal roles not only in transcriptional regulation but also in DNA damage repair processes. CBP/p300 acetyltransferases are considered pivotal transcriptional coactivators in transcriptional regulation processes (Robert G. Roeder, 2019).
Lysine deacetylases
KDACs can be classified into two families based on cofactors facilitating their function: the Zn2+-dependent and NAD+-dependent families. The Zn2+-dependent family, structurally resembling Hda1/Rpd3 proteins, solely exhibits deacetylase activity. Conversely, the NAD+-dependent family, resembling Sir2 proteins, not only serves as lysine deacetylases but also demonstrates ADP-ribosyltransferase activity (Barneda-Zahonero and Parra, 2012).
Methods for Detecting Protein Acetylation
Protein acetylation detection is a crucial technical approach for identifying protein acetylation, acetylation sites, and acetylation levels. Commonly used methods include mass spectrometry, immunoblotting, and chromatin immunoprecipitation assays.
Mass Spectrometry Detection
This method involves the analysis of protein samples using mass spectrometry to identify acetylated proteins and acetylation sites. It offers high sensitivity and specificity, enabling comprehensive analysis of acetylation modifications. However, quantitative analysis of acetylation levels can be relatively challenging. The general workflow includes protein extraction, protein digestion, enrichment of acetylated peptides, mass spectrometry analysis, and data interpretation.
In September 2022, a research paper titled "Acetylome reprogramming participates in the establishment of fruit metabolism during polyploidization in citrus" was published in the Plant Physiology journal. The authors utilized mass spectrometry detection technology to analyze citrus heterozygous tetraploid and parental fruit pulp tissues. The study identified a large number of non-histone proteins with acetylation modifications, revealing over 4,000 acetylation sites in more than 1,600 proteins.
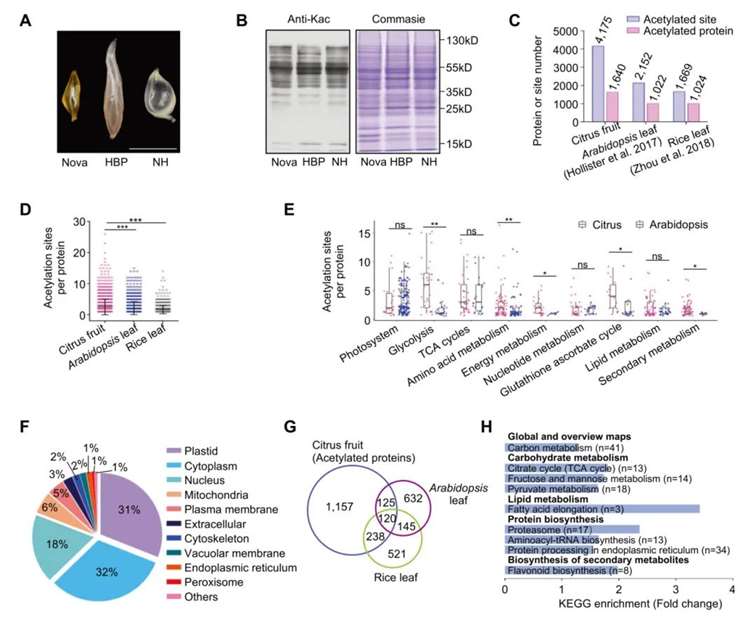 Figure 3 illustrates the investigation of lysine acetyltransferase characteristics in citrus fruits (Zhang et al., 2022). (A) Citrus fruit pulp (juice sacs) used for acetylation analysis, scale bar = 1 cm; (B) Immunoblot analysis of lysine-acetylated proteins in the fruit pulp using anti-acetyllysine (anti-KAc) antibody (left), with Coomassie brilliant blue staining as a control (right); (C) Lysine acetylation (KAc) sites and the quantity of acetylated proteins in citrus, Arabidopsis 5-week-old plant leaves, and rice 14-day-old seedling leaves. Data for Arabidopsis and rice are from Hartl et al. (2017) and Zhou et al. (2018), respectively; (D) Comparison of lysine acetylation sites for each acetylated protein between citrus, Arabidopsis, and rice; (E) Comparison of lysine acetylation sites for each acetylated protein in different metabolic pathways between citrus and Arabidopsis; (F) Predicted subcellular localization distribution of proteins containing acetylated lysine residues in citrus fruits; (G) Venn diagram displaying overlap among homologous acetylated proteins from citrus, Arabidopsis, and rice; (H) KEGG enrichment analysis of citrus-specific acetylated proteins.
Figure 3 illustrates the investigation of lysine acetyltransferase characteristics in citrus fruits (Zhang et al., 2022). (A) Citrus fruit pulp (juice sacs) used for acetylation analysis, scale bar = 1 cm; (B) Immunoblot analysis of lysine-acetylated proteins in the fruit pulp using anti-acetyllysine (anti-KAc) antibody (left), with Coomassie brilliant blue staining as a control (right); (C) Lysine acetylation (KAc) sites and the quantity of acetylated proteins in citrus, Arabidopsis 5-week-old plant leaves, and rice 14-day-old seedling leaves. Data for Arabidopsis and rice are from Hartl et al. (2017) and Zhou et al. (2018), respectively; (D) Comparison of lysine acetylation sites for each acetylated protein between citrus, Arabidopsis, and rice; (E) Comparison of lysine acetylation sites for each acetylated protein in different metabolic pathways between citrus and Arabidopsis; (F) Predicted subcellular localization distribution of proteins containing acetylated lysine residues in citrus fruits; (G) Venn diagram displaying overlap among homologous acetylated proteins from citrus, Arabidopsis, and rice; (H) KEGG enrichment analysis of citrus-specific acetylated proteins.
Immunoblotting Technique
The immunoblotting technique, also known as Western blot (WB), utilizes the principle of specific antigen-antibody binding to detect acetylated proteins. Initially, protein samples undergo separation via polyacrylamide gel electrophoresis. Subsequently, the separated products are transferred onto a solid-phase carrier and bound with a primary antibody, followed by immunoreaction with a specifically labeled secondary antibody. Detection is then achieved through methods such as radiography or chemiluminescence.
In November 2023, a research paper titled "Sir2-mediated cytoplasmic deacetylation facilitates pathogenic fungi infection in host plants" was published in the New Phytologist journal. This study elucidated a novel mechanism by which Fusarium oxysporum f. sp. lycopersici (Fol), a pathogenic fungus, regulates pathogenicity through cytoplasmic protein acetylation. Using subcellular localization and acetylation modification immunoblotting detection, the authors identified a cytoplasm-localized deacetylase, FolSir2, in Fol tomato specialization. Knockout strain ΔFolSir2 exhibited significantly reduced pathogenicity.
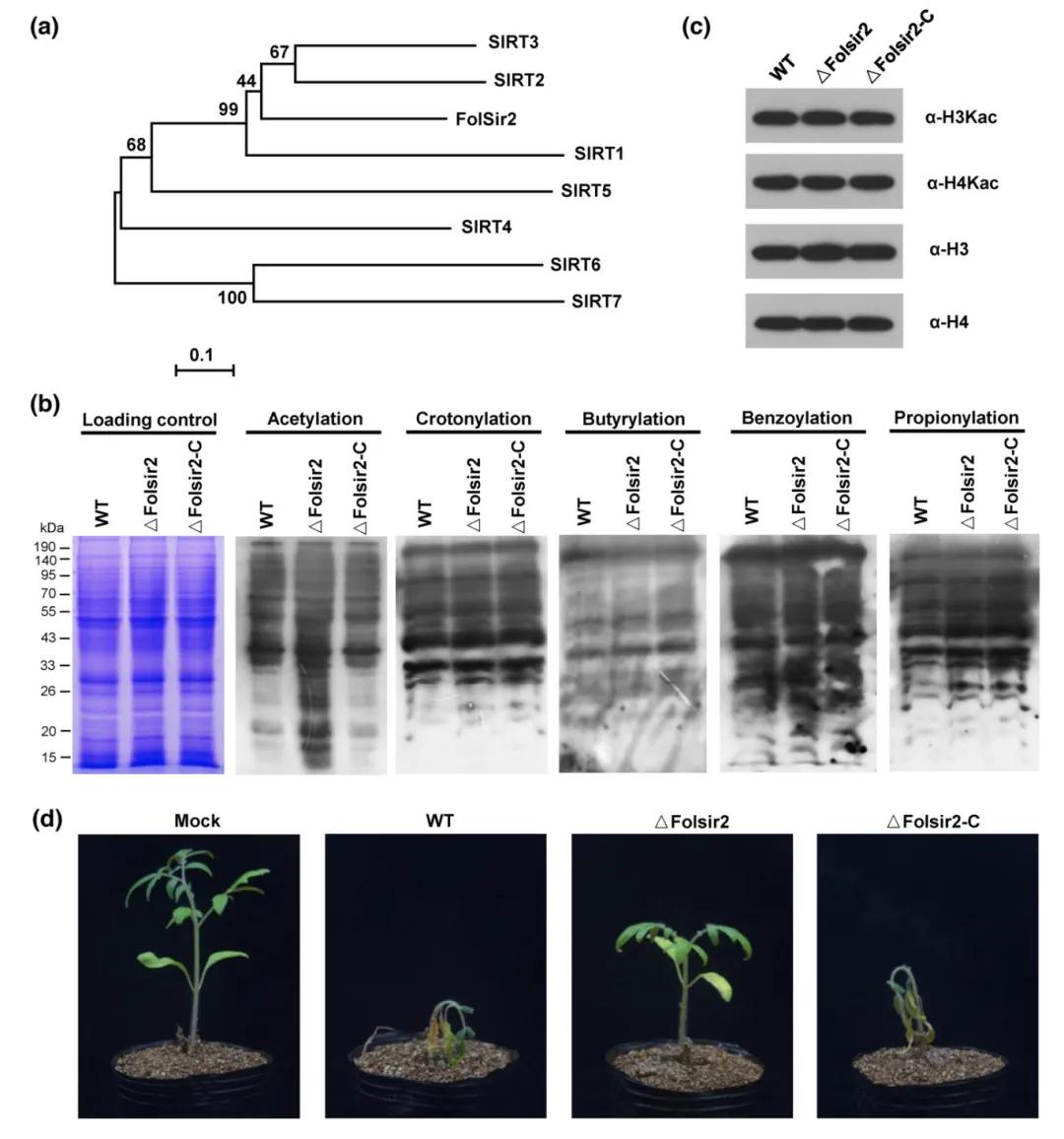 Figure 4 illustrates the critical importance of FolSir2 in deacetylation and pathogenicity of Fusarium oxysporum f. sp. lycopersici (Fol) (Zhang et al., 2023). (a) Establishment of a phylogenetic tree depicting the relationship between FolSir2 and homologous human Sirtuin subtypes (SIRT1-7); (b) Immunoblot analysis of lysine acetylation in wild-type (WT), knockout strain ΔFolSir2, and complemented strain ΔFolSir2-C, with Coomassie brilliant blue staining as a loading control to ensure equal protein loading per lane; each gel shown represents three independent experiments; (c) Immunoblot analysis of histone H3 and H4 acetylation; (d) Assessment of pathogenicity of Fol strains on tomatoes 14 days post-infection.
Figure 4 illustrates the critical importance of FolSir2 in deacetylation and pathogenicity of Fusarium oxysporum f. sp. lycopersici (Fol) (Zhang et al., 2023). (a) Establishment of a phylogenetic tree depicting the relationship between FolSir2 and homologous human Sirtuin subtypes (SIRT1-7); (b) Immunoblot analysis of lysine acetylation in wild-type (WT), knockout strain ΔFolSir2, and complemented strain ΔFolSir2-C, with Coomassie brilliant blue staining as a loading control to ensure equal protein loading per lane; each gel shown represents three independent experiments; (c) Immunoblot analysis of histone H3 and H4 acetylation; (d) Assessment of pathogenicity of Fol strains on tomatoes 14 days post-infection.
Chromatin Immunoprecipitation Assay
The Chromatin Immunoprecipitation (ChIP) technique is a method used to study the interaction between DNA and proteins within a cell. By employing specific antibodies, it selectively enriches particular DNA-binding proteins and their target sites, enabling the investigation of histone modifications and/or protein binding at known target sites within the genome. The experimental procedure typically involves cross-linking, chromatin fragmentation, immunoprecipitation, de-crosslinking, DNA extraction, and data analysis.
In May 2022, a research paper titled "A secreted fungal effector suppresses rice immunity through host histone hypoacetylation" was published in the Nucleic Acids Research journal. This study unveiled a novel mechanism by which the effector protein of Magnaporthe oryzae manipulates rice histone deacetylation to suppress host immunity at the epigenetic level. Employing ChIP-seq analysis, the authors identified a significant reduction in the number of target genes regulated by H3K9 acetylation modification in 35S-UvSec117 transgenic rice. Additionally, RT-qPCR and ChIP-qPCR analyses demonstrated a marked decrease in the expression levels of disease-resistant genes regulated by H3K9 acetylation modification in 35S-UvSec117 transgenic rice.
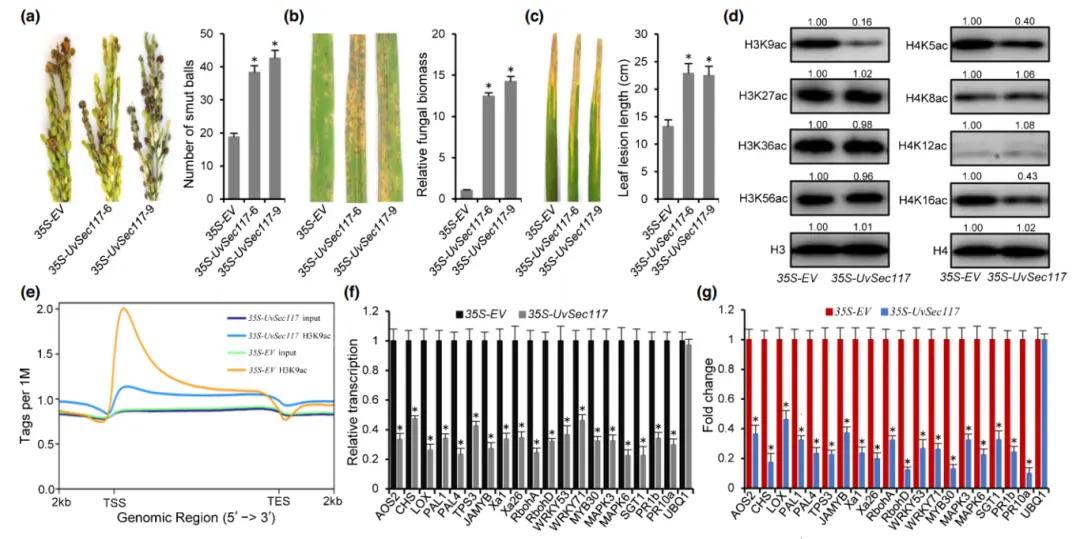 Figure 5: Manipulation of Rice Histone Deacetylase OsHDA701 by the Magnaporthe oryzae Effector Protein UvSec117 Suppresses Rice Immunity (Chen et al., 2022).
Figure 5: Manipulation of Rice Histone Deacetylase OsHDA701 by the Magnaporthe oryzae Effector Protein UvSec117 Suppresses Rice Immunity (Chen et al., 2022).
(a) Left panel: Resistance assessment of 35S-EV and 35S-UvSec117 transgenic lines against Magnaporthe oryzae strain HWD-2 at 25 days post-inoculation. Right panel: Average quantity of Magnaporthe oryzae measured during resistance assessment.
(b) Left panel: Disease symptoms on leaves of 35S-EV and 35S-UvSec117 transgenic lines infected with Magnaporthe oryzae at 11 days post-inoculation. Right panel: Relative fungal biomass of Magnaporthe oryzae strain Pot2 measured by RT-qPCR.
(c) Left panel: Leaf symptoms and length of 35S-EV and 35S-UvSec117 transgenic lines inoculated with Xanthomonas oryzae pv. oryzae strain PXO99A at 21 days post-inoculation (right panel).
(d) Immunoblot analysis of histone acetylation levels (H3K9ac, H3K27ac, H3K36ac, H3K56ac, H4K5ac, H4K8ac, H4K12ac, and H4K16ac) in 35S-EV and 35S-UvSec117 transgenic lines.
(e) Identification of H3K9ac peaks at transcription start sites (TSS), with statistical overlap distribution of read counts in the 2 kb upstream of TSS and 2 kb downstream of transcription end sites (TES) DNA regions.
(f) RT-qPCR analysis of defense-related genes in 35S-EV and 35S-UvSec117 transgenic lines 1 day after Magnaporthe oryzae strain HWD-2 inoculation.
(g) ChIP-qPCR analysis of defense-related genes in 35S-EV and 35S-UvSec117 transgenic lines 1 day after pathogen inoculation.
Protein acetylation, as a crucial post-translational modification, plays a pivotal role in cellular function regulation and signal transduction processes. To delve deeper into the functionality and mechanistic actions of protein acetylation, a diverse array of research methodologies can be employed. While the previous discussion focused on elucidating specific techniques, an overarching view reveals the integrated application of various research approaches by the authors, which may further interest readers inclined to explore independently.
The aforementioned three methods are currently widely utilized in the field of research. However, with the rapid advancements in high-throughput proteomics, chemical biology, and bioinformatics, we anticipate obtaining a more profound and comprehensive understanding of protein acetylation modifications. The future research outlook is brimming with boundless anticipation.
The Functional Role of Protein Acetylation Modification in Plants
Protein acetylation modification plays a crucial role in vital physiological processes and disease-related mechanisms in organisms, including but not limited to the regulation of gene transcription, activation of DNA damage repair mechanisms, precise execution of cell division, modulation of signal transduction pathways, facilitation of protein folding processes, regulation of autophagic activities, and maintenance of metabolic balance. However, what specific functions does protein acetylation modification confer at the individual plant level?
Involvement in Plant Immune Response
The interaction mechanisms between pathogen effector proteins and plants have long been a focal point in plant research. However, there remain unresolved questions regarding how these effectors activate plant immunity. A research paper titled "Direct acetylation of a conserved threonine of RIN4 by the bacterial effector HopZ5 or AvrBsT activates RPM1-dependent immunity in Arabidopsis" was published in the Molecular Plant journal. This study reveals a novel mechanism by which the pathogen effector proteins HopZ5 and AvrBsT activate plant immunity through the acetylation modification of RIN4 (RPM1-INTERACTING PROTEIN4).
Previous studies have indicated that the Arabidopsis deacetylase SOBER1 can suppress the immune response triggered by the effector proteins HopZ5 and AvrBsT. In this study, the authors found that in the absence of SOBER1, the immune response activated by these two pathogen effector proteins depends on RIN4 and the intracellular resistance protein RPM1. The authors performed a dependency validation of acetyltransferase effector protein-triggered hypersensitivity reaction (HR) using the Arabidopsis SOBER1 loss-of-function mutant sober1-3, abbreviated as sober1. The growth of Pto DC3000 pathogens carrying the effector proteins HopZ5 or AvrBsT in the sober1 rpm1 and sober1 rps2 rin4 mutants is comparable to that of strains carrying an empty vector of Pto DC3000, but differs from that in the sober1 rps2 mutant, indicating that in Arabidopsis, the immune response triggered by HopZ5 or AvrBsT requires RPM1 and RIN4, rather than RPS2. In conclusion, bacterial acetyltransferases HopZ5 and AvrBsT may target RIN4 to activate RPM1-dependent immunity.
 Figure 6 Bacterial acetyltransferase effector proteins HopZ5 and AvrBsT trigger RPM1- and RIN4-dependent immunity in Arabidopsis (Choi et al., 2021).
Figure 6 Bacterial acetyltransferase effector proteins HopZ5 and AvrBsT trigger RPM1- and RIN4-dependent immunity in Arabidopsis (Choi et al., 2021).
Other studies have shown that two additional effector proteins, AvrB and AvrRpm1, can induce the phosphorylation of threonine 166 (T166) of RIN4 and activate the RPM1-dependent immune response through the cytoplasmic receptor kinase RIPK. Therefore, the interaction between HopZ5 and AvrBsT with RIN4 was further investigated by the authors.
Through co-immunoprecipitation experiments, researchers found a direct interaction between HopZ5 and AvrBsT with RIN4 (Figure 7A). Interestingly, through in vitro assays and mass spectrometry identification, it was discovered that as acetyltransferases, HopZ5 and AvrBsT can acetylate the conserved site T166 of RIN4 (Figure 7B, C). This acetylation modification not only occurs in vitro but also in planta and during pathogen invasion (Figure 7D, E). Ultimately, acetylation modification of the T166 site on RIN4 activates the RPM1-dependent immune response, enabling plants to resist pathogen invasion and exhibit disease resistance.
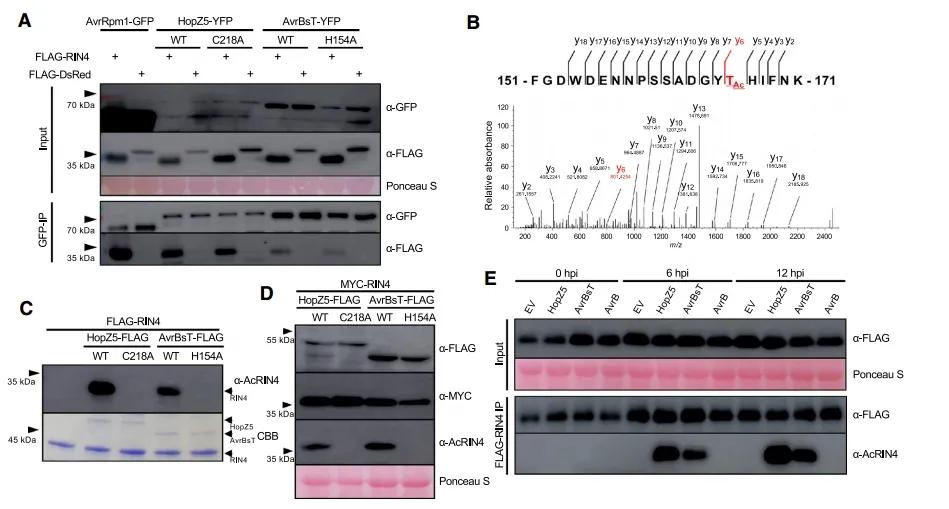 Figure 7 depicts the interaction and acetylation of RIN4 by HopZ5 and AvrBsT (Choi et al., 2021). (A) Interaction between HopZ5 and AvrBsT with RIN4, regardless of their catalytic activity. (B) MS/MS spectrum of the RIN4 peptide fragment (FGDWDENNPSSADGYTHIFNK) acetylated after incubation with HopZ5-FLAG protein. (C) In vitro studies of HopZ5 and AvrBsT acetylating RIN4. (D) Acetylation of the T166 site of RIN4 by HopZ5 and AvrBsT in N. benthamiana. (E) Acetylation of T166 RIN4 by HopZ5 in Arabidopsis thaliana.
Figure 7 depicts the interaction and acetylation of RIN4 by HopZ5 and AvrBsT (Choi et al., 2021). (A) Interaction between HopZ5 and AvrBsT with RIN4, regardless of their catalytic activity. (B) MS/MS spectrum of the RIN4 peptide fragment (FGDWDENNPSSADGYTHIFNK) acetylated after incubation with HopZ5-FLAG protein. (C) In vitro studies of HopZ5 and AvrBsT acetylating RIN4. (D) Acetylation of the T166 site of RIN4 by HopZ5 and AvrBsT in N. benthamiana. (E) Acetylation of T166 RIN4 by HopZ5 in Arabidopsis thaliana.
The findings of this investigation demonstrate that HopZ5 and AvrBsT activate the RPM1-dependent immune response by directly acetylating the RIN4 protein. This unveils a novel mechanism whereby immune responses in plants are regulated through acetylation modification. The discoveries of this study hold significant implications for a comprehensive understanding of the interaction between plants and pathogenic microorganisms, elucidating the mechanisms of immune signal transduction, and the development of disease-resistant technologies.
Participation in Plant Stress Response
Protein acetylation plays a crucial regulatory role in plant responses to adverse environmental conditions by modulating protein activity, stability, and gene expression, thereby aiding plants in adapting to stress environments and enhancing stress resistance. Investigating the relationship between protein acetylation and plant stress response contributes to our comprehension of plant stress adaptation mechanisms and provides a theoretical foundation for breeding stress-tolerant crops.
In January 2022, the journal Stress Biology published a research article titled "Acetylproteomics analyses reveal critical features of lysine-ε-acetylation in Arabidopsis and a role of 14-3-3 protein acetylation in alkaline response," which elucidated protein acetylation in major organs of Arabidopsis, analyzed key features of lysine acetylation in plants, and unveiled the significant role of 14-3-3 protein acetylation in plant alkaline stress response.
In this study, high-resolution tandem mass spectrometry was employed by the authors to conduct lysine acetylation analysis on five representative plant organs of Arabidopsis, including leaves, roots, stems, flowers, and seeds. A total of 2887 Kac (Lysine-ε-acetylation) proteins and 5929 Kac sites were identified. Evolutionary analysis revealed that Kac predominantly targets evolutionarily conserved proteins and conserved lysine residues.
The authors observed that many Kac proteins are also regulated by other post-translational modifications (such as phosphorylation, ubiquitination, SUMOylation). Although acetylation, ubiquitination, and SUMOylation can all modify lysine residues, they seldom occur at the same sites. Proteins targeted by both acetylation and phosphorylation typically contain numerous bromo domains and 14-3-3 protein domains. Structural analysis indicated that acetylation of lysine 56 (K56ac) at the binding site of 14-3-3 proteins to phosphorylated substrates may impede their interaction with phosphorylated substrates (Figure 8A). Previous studies have demonstrated that phosphorylation recognition protein 14-3-3s participate in the regulation of plasma membrane H+-ATPase AHA2 activity, and the interaction between 14-3-3s and AHA2 is regulated by post-translational modifications of 14-3-3s proteins themselves.
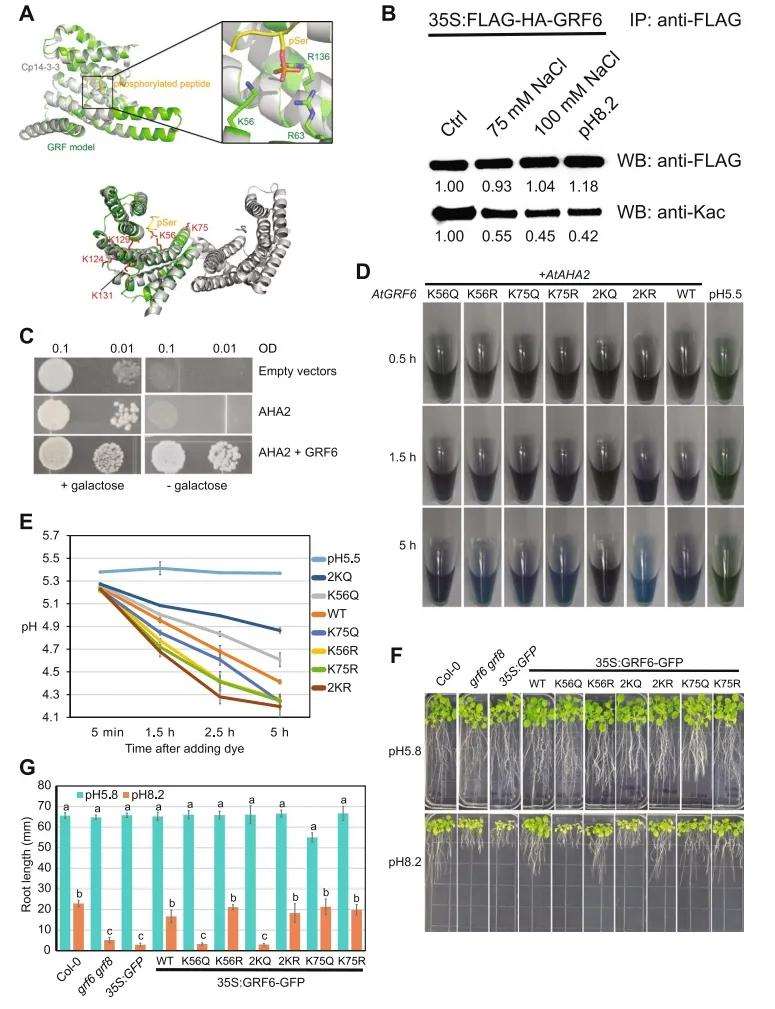 Figure 8 illustrates the negative regulation of Arabidopsis alkalinity tolerance by Lys56 acetylation in GRF6 (Guo et al., 2022). (A) The predicted model of GRF6 based on Cp14-3-3 reveals its structure. The enlarged view (top) indicates the phosphorylated serine (pSer) binding pocket containing three positively charged residues (K56, R63, and R136), with the side chain of acetylated lysine marked in red (bottom). (B) Immunoprecipitation and immunoblot analysis of GRF6 protein and acetylation levels. (C) Yeast assay demonstrates the requirement of both AtGRF6 and AtAHA2 for yeast growth when glucose is the sole carbon source. Growth of yeast is independent of AtGRF6 and AHA2 when galactose is the sole carbon source (left panel). (D) Methyl violet staining shows the impact of AtAHA2 and different AtGRF6 expressions on culture medium pH. (E) Quantification of pH changes in (D). (F) Phenotypes of plant root growth on normal pH 5.8 and high pH 8.2 plates (G) Quantification of root length in (F).
Figure 8 illustrates the negative regulation of Arabidopsis alkalinity tolerance by Lys56 acetylation in GRF6 (Guo et al., 2022). (A) The predicted model of GRF6 based on Cp14-3-3 reveals its structure. The enlarged view (top) indicates the phosphorylated serine (pSer) binding pocket containing three positively charged residues (K56, R63, and R136), with the side chain of acetylated lysine marked in red (bottom). (B) Immunoprecipitation and immunoblot analysis of GRF6 protein and acetylation levels. (C) Yeast assay demonstrates the requirement of both AtGRF6 and AtAHA2 for yeast growth when glucose is the sole carbon source. Growth of yeast is independent of AtGRF6 and AHA2 when galactose is the sole carbon source (left panel). (D) Methyl violet staining shows the impact of AtAHA2 and different AtGRF6 expressions on culture medium pH. (E) Quantification of pH changes in (D). (F) Phenotypes of plant root growth on normal pH 5.8 and high pH 8.2 plates (G) Quantification of root length in (F).
In further exploration of the biological functions of 14-3-3 protein acetylation, a yeast heterologous expression system was developed for validation. Arabidopsis AHA2 and 14-3-3 protein GRF6 were introduced into yeast lacking endogenous 14-3-3 and H+-ATPase proteins, creating acetylation (K56Q) and deacetylation (K56R) mutants (Figure 8C). pH changes in yeast extracellular solutions were monitored, revealing a negative regulatory effect of Lys56 acetylation in GRF6 on AHA2 activity. Transgenic experiments in Arabidopsis also confirmed the negative regulatory role of GRF6 Lys56 acetylation in alkaline stress response (Figures 8D, E). In the grf6 grf8 double mutant background, introduction of GRF6 K56Q failed to restore normal plant growth under high pH conditions, and acetylation levels of GRF6 decreased under high pH conditions (Figures 8F, G). Thus, this study elucidates a novel mechanism whereby protein acetylation regulates plant alkaline stress response by modulating the interaction between 14-3-3 proteins and AHA2.
References
- Aksnes, H., Drazic, A., Marie, M., et al. (2016). First things first: vital protein marks by N-terminal acetyltransferases. Trends in Biochemical Sciences, 41(9), 746-760.
- Nagy, Z., & Tora, L. (2007). Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene, 26(37), 5341-5357.
- Fournier, M., Orpinell, M., Grauffel, C., et al. (2016). KAT2A/KAT2B-targeted acetylome reveals a role for PLK4 acetylation in preventing centrosome amplification. Nature Communications, 7(1), 13227.
- Barneda-Zahonero, B., & Parra, M. (2012). Histone deacetylases and cancer. Molecular Oncology, 6(6), 579-589.
- Zhang, M., Tan, F. Q., Fan, Y. J., et al. (2022). Acetylome reprograming participates in the establishment of fruit metabolism during polyploidization in citrus. Plant Physiology, 190(4), 2519-2538.
- Zhang, N., Hu, J., Liu, Z., et al. (2023). Sir2‐mediated cytoplasmic deacetylation facilitates pathogenic fungi infection in host plants. New Phytologist.
- Choi, S., Prokchorchik, M., Lee, H., et al. (2021). Direct acetylation of a conserved threonine of RIN4 by the bacterial effector HopZ5 or AvrBsT activates RPM1-dependent immunity in Arabidopsis. Molecular Plant, 14(11), 1951-1960.
- Guo, J., Chai, X., Mei, Y., et al. (2022). Acetylproteomics analyses reveal critical features of lysine-ε-acetylation in Arabidopsis and a role of 14-3-3 protein acetylation in alkaline response. Stress Biology, 2(1), 1.
- Wang, R., Sun, H., Wang, G., et al. (2020). Imbalance of lysine acetylation contributes to the pathogenesis of Parkinson's disease. International Journal of Molecular Sciences, 21(19), 7182.
- Roeder, R. G. (2019). 50+ years of eukaryotic transcription: an expanding universe of factors and mechanisms. Nature Structural & Molecular Biology, 26(9), 783-791.
- Chen, X., Duan, Y., Qiao, F., et al. (2022). A secreted fungal effector suppresses rice immunity through host histone hypoacetylation. New Phytologist, 235(5), 1977-1994.
Our products and services are for research use only.