Protein phosphorylation is a vital post-translational modification (PTM) that plays a key role in regulating numerous cellular processes, such as signal transduction, cell cycle progression, and metabolic control. This modification involves the addition of a phosphate group to specific amino acids—primarily serine, threonine, or tyrosine—within proteins. This addition can significantly change the protein's structure and function, influencing how it interacts with other molecules in the cell.
To delve deeper into this fascinating modification, check out our resource: What is Protein Phosphorylation?
What is Protein Phosphorylation Site Prediction
Phosphorylation site prediction refers to the use of computational tools designed to identify potential phosphorylation sites within protein sequences. Accurate predictions are crucial for gaining insights into protein function and regulatory mechanisms, as well as for developing targeted therapeutic strategies.
Why Use Phosphorylation Site Prediction Tools?
Employing computational tools for predicting phosphorylation sites comes with several advantages:
- Efficiency: These tools can swiftly pinpoint potential phosphorylation sites, saving researchers from extensive laboratory experiments.
- Cost-Effectiveness: By reducing reliance on expensive experimental methods, these tools help lower research costs.
- Enhanced Accuracy: Advanced algorithms analyze sequence patterns and functional features, significantly improving prediction accuracy.
- In summary, leveraging phosphorylation site prediction tools not only streamlines research but also enhances our understanding of protein dynamics in biological systems.
Workflow of Prediction of Phosphorylation Sites
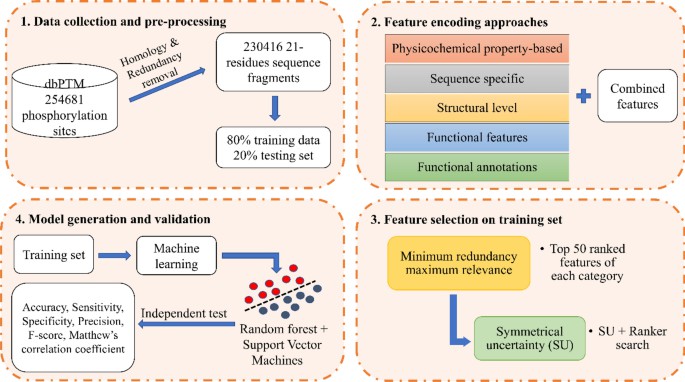 Figure 1 Overall workflow of the proposed approach for prediction of phosphorylation sites (Salma Jamal et al,. 2021)
Figure 1 Overall workflow of the proposed approach for prediction of phosphorylation sites (Salma Jamal et al,. 2021)
Select Service
Tools for Phosphorylation Site Prediction
Protein phosphorylation is a crucial post-translational modification that regulates various cellular processes. Accurate prediction of phosphorylation sites is essential for understanding protein function and developing targeted therapeutic strategies. Several powerful tools are available to assist researchers in predicting these sites, each with unique features and methodologies.
Overview of Popular Phosphorylation Site Prediction Tools
Here are some of the most notable tools for predicting phosphorylation sites:
1. PhosphoPredict
PhosphoPredict combines sequence and functional features to predict kinase-specific substrates and their associated phosphorylation sites for various human kinases.
Key Features:
Utilizes a comprehensive dataset from multiple databases.
Competitive performance compared to other tools, achieving high accuracy in predictions.
Accuracy: Demonstrated competitive performance with over 99% specificity for the human proteome (Jiang et al., 2016) .
2. KinasePhos
KinasePhos specializes in kinase-specific predictions using machine learning algorithms.
Key Features:
Utilizes a large dataset of experimentally verified phosphorylation sites.
Provides predictive models for individual kinases, families, and groups.
Accuracy: Achieved accuracies of 94.5% for protein kinase B and around 87.2% on average across all models (Zhang et al., 2022) .
3. DeepPhos
DeepPhos employs a deep learning architecture to predict phosphorylation sites.
Key Features:
Uses densely connected convolutional neural networks to capture complex sequence representations.
Capable of kinase-specific predictions at various levels (group, family, individual).
Performance: Outperforms many existing methods in both general and kinase-specific predictions (Liu et al., 2019) .
4. Attenphos
Attenphos is based on a self-attention mechanism designed to enhance prediction accuracy.
Key Features:
Captures long-range dependencies between amino acids effectively.
Reduces model parameters while improving efficiency and generalization.
Performance: Demonstrated superior performance in predicting serine, threonine, and tyrosine phosphorylation sites compared to other state-of-the-art methods (Gao et al., 2024).
5. TransPhos
TransPhos utilizes a transformer encoder along with densely connected convolutional neural network blocks for prediction.
Key Features:
Focuses on improving prediction accuracy through advanced deep learning techniques.
Performs well on datasets of serine, threonine, and tyrosine phosphorylation sites.
Performance: Achieved high AUC values across various tests, outperforming several established tools (Liu et al., 2022).
6. RF-Phos: Random Forest-Based Phosphosite Predictor
RF-Phos utilizes a random forest algorithm primarily to predict phosphorylation sites based on the primary amino acid sequence of proteins. This tool is designed to identify potential phosphorylation sites without relying on complex structural information or additional functional data.
Key Features:
Simplicity: RF-Phos does not depend on intricate structural data, making it accessible for a wider range of applications.
Pattern Recognition: It effectively captures complex patterns surrounding phosphorylation residues, allowing for more accurate predictions.
Accuracy: RF-Phos has demonstrated higher accuracy compared to other prediction methods, making it a reliable choice for researchers.
7. PhosIDN: Integrated Deep Neural Network
The PhosIDN model integrates sequence and protein-protein interaction (PPI) information for improved phosphorylation site prediction. This approach significantly enhances prediction performance compared to existing methods by effectively combining diverse features.
Key Features:
Data Integration: By combining multiple data sources, PhosIDN provides more comprehensive and accurate predictions.
Performance Improvement: The model shows significant improvements in prediction performance, especially when dealing with phosphorylation sites that have complex biological contexts.
8. PPRED: A Generalized Prediction System
PPRED is a generalized phosphorylation site prediction system that relies on evolutionary information rather than kinase-specific data for its predictions.
Key Features:
Evolutionary Conservation: Utilizes conserved features from protein sequences to classify potential phosphorylation sites.
Broad Applicability: Suitable for proteins without known kinase data, making it versatile in various research contexts.
In a study by Ashis et al. (2010), the PPRED system was developed to predict phosphorylation sites using evolutionary information without relying on kinase-specific data. The results indicated that PPRED achieved an accuracy of approximately 68% for predicting phospho-serine sites, demonstrating its effectiveness in classifying phosphorylation sites based solely on protein sequence data.
Table: Comparison of Popular Tools
| Tool Name | Key Features | Accuracy Rate |
|---|---|---|
| PhosphoPredict | Combines sequence and functional features | >99% specificity |
| KinasePhos | Machine learning-based with high specificity | Up to 94.5% |
| DeepPhos | Deep learning architecture for enhanced prediction | Superior to traditional methods |
| Attenphos | Self-attention mechanism for improved accuracy | Best overall performance in recent tests |
| TransPhos | Transformer encoder with dense connections | High AUC values |
| RF-Phos | Random forest-based; captures complex patterns in sequences | High accuracy compared to traditional methods |
| PhosIDN | Integrated deep neural network; incorporates PPI information | Enhanced prediction performance |
| PPRED | Uses evolutionary data; independent of kinase specificity | ~68% accuracy for phospho-serine sites |
The availability of various phosphorylation site prediction tools has significantly advanced our understanding of protein functions and regulatory mechanisms. Each tool offers unique features tailored to specific research needs, making it essential for researchers to choose the most appropriate tool based on their specific requirements.
Key Features of Effective Prediction Tools
When it comes to predicting phosphorylation sites in proteins, the effectiveness of a prediction tool is crucial for obtaining accurate and reliable results. Here are some key features that make these tools effective:
1. High-Quality Datasets
Well-Annotated Data: Effective prediction tools rely on high-quality datasets that are well-annotated with experimentally verified phosphorylation sites. This ensures that the model is trained on accurate information, which improves the reliability of predictions.
Extensive Training Data: A larger dataset allows the tool to learn from a wider variety of examples, enhancing its ability to generalize and make accurate predictions across different proteins and conditions.
2. Advanced Algorithms
Machine Learning Capabilities: Tools that utilize machine learning algorithms can analyze complex patterns within protein sequences more effectively than traditional methods. These algorithms can adapt and improve over time as more data becomes available.
Artificial Intelligence Integration: Incorporating AI techniques, such as deep learning, allows for the development of models that can capture intricate relationships between amino acids and their phosphorylation status, leading to higher prediction accuracy.
3. Functional Integration
Insights from Protein-Protein Interactions: Tools that integrate information about protein-protein interactions can provide context for how phosphorylation affects protein function. Understanding these interactions is vital for predicting the biological significance of phosphorylation sites.
Cellular Pathway Context: Incorporating knowledge of cellular pathways helps in understanding the functional implications of phosphorylation. This integration allows researchers to make more informed predictions about how specific modifications may influence cellular processes.
Machine Learning in Phosphorylation Prediction
Machine learning has transformed phosphorylation site prediction by enabling pattern recognition in vast datasets. Methods like Random Forests and Support Vector Machines improve predictive accuracy, while deep learning methods such as convolutional neural networks (CNNs) have further advanced the field.
Explore related detection methods here: Protein Phosphorylation Detection Methods.
Phosphorylation Prediction Databases
Phosphorylation prediction databases are vital resources for researchers aiming to understand phosphorylation sites and their implications in cellular processes. These databases compile extensive data on phosphorylation events, enabling scientists to make informed decisions in their research. Below are some notable databases that provide valuable information regarding phosphorylation sites.
1. PhosphoSitePlus
PhosphoSitePlus is a comprehensive database that focuses on post-translational modifications (PTMs), particularly phosphorylation. It provides detailed information about phosphorylation sites, including experimental evidence and associated kinases.
Key Features:
- Extensive collection of over 130,000 non-redundant phosphorylation sites.
- User-friendly interface for searching and analyzing data.
Hornbeck et al. (2015) demonstrated that PhosphoSitePlus serves as a critical resource for identifying potential biomarkers in cancer research by providing a wealth of information on phosphorylation events associated with various signaling pathways.
2. UniProt
UniProt is a leading protein sequence database that offers detailed annotations on protein functions, including post-translational modifications such as phosphorylation.
Key Features:
- High-quality annotations based on experimental data.
- Comprehensive coverage of proteins across various organisms.
The UniProt database has been instrumental in advancing our understanding of protein functions and interactions through its extensive annotation system, as highlighted by The UniProt Consortium (2021).
3. PPSP (Phosphorylation Site Prediction)
PPSP is a focused database dedicated to predicting phosphorylation sites using various algorithms to enhance prediction accuracy.
Key Features:
- Integration of multiple prediction algorithms.
- User-friendly interface for inputting sequences and obtaining predictions.
Zhao et al. (2013) showcased the effectiveness of PPSP in predicting kinase-specific phosphorylation sites, emphasizing its role in understanding kinase-substrate interactions.
4. PhosphoNetworks
PhosphoNetworks is a database that provides a high-resolution map of human phosphorylation networks, detailing kinase-substrate relationships and specific phosphorylation sites.
Key Features:
- Comprehensive dataset of kinase-substrate relationships from high-throughput experiments.
- Analytical tools for dissecting phosphorylation networks.
A study by Keshava et al. (2013) highlighted the utility of PhosphoNetworks in exploring the complexities of cellular signaling pathways by mapping kinase-substrate interactions at specific phosphorylation sites.
5. PHOSIDA
PHOSIDA is a comprehensive database that integrates thousands of phosphosites identified through mass spectrometry across various species.
Key Features:
- High-confidence phosphosite data integrated from large-scale proteomics studies.
- Predictive capabilities based on machine learning techniques.
Research by Schaeffer et al. (2007) demonstrated the effectiveness of PHOSIDA in retrieving and analyzing phosphosites from quantitative phosphoproteomics experiments, highlighting its role in understanding biological responses to stimuli.
6. dbPAF
dbPAF is an integrative database that compiles extensive data on protein phosphorylation across various species, including humans and model organisms.
Key Features:
- Contains over 54,000 phosphoproteins with nearly half a million phosphorylation sites.
- Tools for motif detection and potential kinase prediction based on collected phospho-site data.
A study by Zhang et al. (2016) utilized dbPAF to analyze evolutionary conservation states across different species, demonstrating its utility in comparative studies of protein phosphorylation.
7. GPS 6.0
- An updated server that predicts kinase-specific phosphorylation sites using advanced machine learning techniques on a large dataset of non-redundant phosphosites
8. CKSAAP_PhSite
- An online tool that predicts phosphorylation sites based solely on sequence information, achieving impressive sensitivity and specificity rates for different amino acids
9. RF-Phos 2.0:
- Utilizes random forest algorithms to predict phosphorylation sites based on primary amino acid sequences, demonstrating high accuracy compared to other methods
Table: Comparison of Major Databases
| Database | Description | Key Features |
|---|---|---|
| PhosphoSitePlus | Comprehensive resource for PTMs | Extensive datasets; user-friendly interface |
| UniProt | Detailed protein sequence annotations | High-quality annotations; broad coverage |
| PPSP | Focused database for phosphorylation prediction | Multiple algorithms; easy-to-use interface |
| PhosphoNetworks | High-resolution map of phosphorylation networks | Kinase-substrate relationships; analytical tools |
| PHOSIDA | Integrates thousands of phosphosites | High-confidence data; predictive capabilities |
| dbPAF | Integrative database for protein phosphorylation | Large dataset; motif detection tools |
| GPS 6.0 | Updated server for kinase-specific phosphorylation sites | Utilizes advanced machine learning on a large dataset of non-redundant phosphosites; high accuracy in predictions |
| CKSAAP_PhSite | Online tool predicting phosphorylation sites based on sequence information | Achieves high sensitivity and specificity rates for different amino acids |
| RF-Phos 2.0 | Predicts phosphorylation sites using random forest algorithms | Demonstrates high accuracy compared to other methods |
Applications of Prediction Tools in Research
Phosphorylation site prediction tools are essential in various fields of biological research, including drug discovery, disease research, and the study of cellular signaling pathways. Below are detailed examples that illustrate these applications.
Drug Discovery: Identifying Therapeutic Targets
Phosphorylation plays a critical role in cellular signaling and is often implicated in disease mechanisms. Tools like PhosphoPredict have been developed to predict human kinase-specific phosphorylation sites, which can aid in identifying potential therapeutic targets. For instance, PhosphoPredict integrates protein sequence and functional features to predict substrates for multiple kinases, significantly enhancing the identification of kinase-specific phosphorylation sites across the human proteome.
This capability is crucial for drug discovery as it allows researchers to pinpoint specific phosphorylation sites that may be targeted by new therapeutics.
Disease Research: Elucidating Mechanisms of Conditions
Phosphorylation site prediction tools are also pivotal in understanding diseases such as cancer and diabetes. For example, the study by Laukens et al. utilized conditional random fields (CRF) to predict phosphorylation sites, demonstrating that this method outperforms existing techniques when applied to experimentally verified data sets. Such predictive models can illuminate the underlying mechanisms of disease by identifying key phosphorylation events that drive pathological processes.
Cellular Signaling Pathways: Understanding Complex Biological Networks
Understanding cellular signaling pathways is another significant application of phosphorylation site prediction tools. The development of tools like DeepPhos, which employs deep learning architectures for predicting phosphorylation sites, illustrates advancements in this area. DeepPhos has shown superior performance compared to traditional methods by effectively leveraging complex data representations to enhance prediction accuracy. This capability is vital for mapping out intricate signaling networks and understanding how various pathways interact within a cell.
The integration of advanced computational tools for phosphorylation site prediction is transforming our understanding of biological processes and diseases. These tools not only facilitate drug discovery but also provide insights into disease mechanisms and cellular signaling pathways.
Discover how PTMs impact proteins in our detailed guide: What is Post-Translational Modifications (PTMs)?.
Challenges and Future Directions in Protein Phosphorylation Research
Phosphorylation site prediction faces several key challenges that hinder its full potential:
- Limited data availability: Particularly for proteins that remain under-researched, making it difficult to create comprehensive models.
- Kinase specificity: The variability in kinase-target interactions complicates predictions, as different kinases exhibit unique preferences.
- Computational complexity: The demand for high-throughput and scalable analysis methods grows with increasing data volumes.
Addressing these challenges will require innovations in both data collection methods and algorithm development. Emerging technologies hold the promise to overcome these obstacles and drive future advancements:
- CRISPR-based screenings: These tools allow for precise functional studies, uncovering previously unknown phosphorylation pathways.
- Advanced mass spectrometry: This technique is transforming our understanding of phosphorylation dynamics with unparalleled detail and accuracy.
- Artificial intelligence (AI): By leveraging AI, prediction models are becoming more accurate and efficient, while complex datasets are being interpreted with greater precision.
Together, these advancements pave the way for more robust and insightful research in protein phosphorylation, bridging current gaps and opening new avenues for scientific discovery.
Conclusion: Why Accurate Predictions Matter
Protein phosphorylation site prediction is essential for advancing biological research and therapeutic development. Accurate tools not only accelerate discovery but also contribute to a deeper understanding of cellular mechanisms.
Take Action Now: Partner with Creative Proteomics to leverage our cutting-edge phosphorylation analysis services and accelerate your research.
Relevant Questions People Also Ask
What are the best tools for predicting protein phosphorylation sites?
Some of the leading tools include PhosphoPredict, KinasePhos, and DeepPhos, each utilizing different methodologies such as machine learning and sequence analysis to enhance prediction accuracy.
How accurate are phosphorylation site prediction tools?
The accuracy varies by tool, but recent studies show that advanced methods like Attenphos demonstrate significantly higher accuracy rates compared to traditional approaches, often exceeding 90% in specific datasets .
What factors influence the prediction of phosphorylation sites?
Key factors include the choice of algorithm (e.g., machine learning vs. consensus patterns), the quality and size of training datasets, and the incorporation of functional features such as protein-protein interactions .
Can these tools predict kinase-specific phosphorylation sites?
Yes, many tools like GPS and Musite are designed to predict kinase-specific sites by analyzing sequence motifs and structural features associated with specific kinases.
What are the applications of phosphorylation site prediction?
Applications include drug discovery, understanding disease mechanisms, and elucidating cellular signaling pathways.
References
- Jiang, L., Chen, H., Pinello, L., & Yuan, G.-C. (2016). GiniClust: Detecting rare cell types from single-cell gene expression data using the Gini index. Genome Biology, 17, article 144. https://doi.org/10.1186/s13059-016-1010-4.
- Laukens, K., et al. (2008). Prediction of kinase-specific phosphorylation sites using conditional random fields. Bioinformatics, 24(24), 2857-2864. https://doi.org/10.1093/bioinformatics/btn501.
- Gao, X., et al. (2019). DeepPhos: prediction of protein phosphorylation sites with deep learning. Bioinformatics, 35(16), 2766-2775. https://doi.org/10.1093/bioinformatics/btz1112.
- Huang, Y., et al. (2015). Musite: a web server for predicting protein phosphorylation sites based on protein disorder scores and local amino acid sequence frequencies. Bioinformatics, 31(12), 1977-1979. https://doi.org/10.1093/bioinformatics/btv042.
Our products and services are for research use only.

