The role of proteolytic enzymes in cellular function regulation has long been underestimated by the scientific realm. The identification of ubiquitin and its associated proteolytic enzymes has marked a revolutionary shift in scientific insights. Ubiquitin, a highly conserved molecule consisting of 76 amino acids, is ubiquitous in eukaryotic cells – hence its denomination. Its covalent conjugation with proteins enables them to be recognized and degraded by proteases, offering a universal pathway for degrading short-lived and aberrant proteins within the cell. Contrasting the protein hydrolysis occurring in the digestive tract, the ubiquitin-protein hydrolysis process, from the binding of ubiquitin to a protein through to its degradation into smaller peptide segments, necessitates energy participation. Scientists have begun to recognize the importance of the ubiquitin-protease system as a critical regulatory mechanism in eukaryotes.
Select Service
Ubiquitin-Protease System
Ubiquitin-protease system is a regulatory mechanism found in all eukaryotic cells. Between the 1970s to 1980s, the enigma of how ubiquitin mediates protein degradation was unravelled (Figure 1). This degradation process necessitates the involvement of three enzymes: ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2), and ubiquitin protein ligase (E3). The E3 enzyme is responsible for the specific recognition of the proteins being degraded during the ubiquitination process. The ubiquitination process mediated by E2s and E3s can be reversed by deubiquitinating enzymes (DUBs). Currently identified DUBs can be categorised into two main types: ubiquitin C-terminal hydrolases (UCHs) and ubiquitin-specific processing proteases (UBPs), both of which are cysteine proteases. Typically, UCHs mainly hydrolyse ester and ubiquitin's amino linkage at the carbonyl end and can also decompose ubiquitin precursors to generate active ubiquitin molecules; UBPs degrade ubiquitin polymer chains.
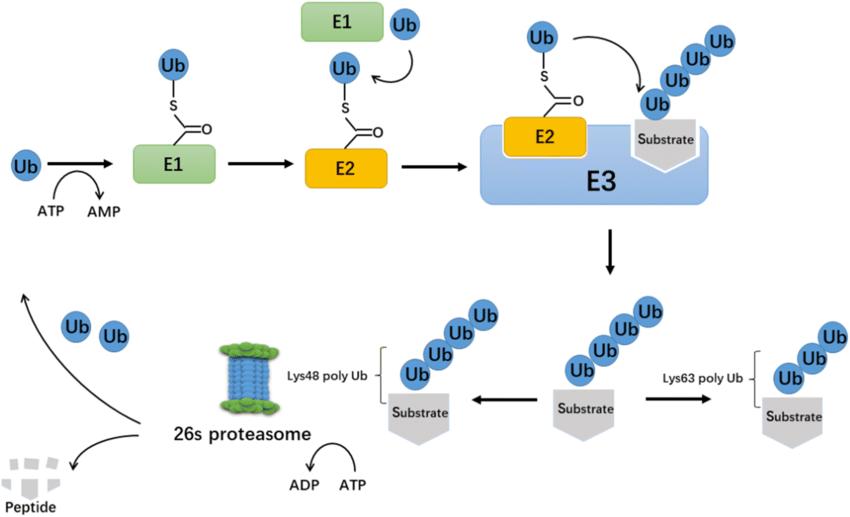 The process of ubiquitination. (Xiaofeng Gong et al,. 2020)
The process of ubiquitination. (Xiaofeng Gong et al,. 2020)
Ubiquitination in physiological processes
Ubiquitination plays a crucial role in physiological processes such as cell differentiation, organelle biosynthesis, apoptosis, DNA repair, new protein generation, cell proliferation regulation, protein transport, immune response, and stress response. Bence et al. found that protein deposition could directly weaken the function of the ubiquitin-proteasome system. Consequently, the transient expression of two unrelated but aggregating proteins could almost completely inhibit the ubiquitin-proteasome system. Given the significant role of ubiquitin-mediated protein degradation in regulating cell activities, underlying mechanisms causing protein aggregation could disrupt cellular homeostasis leading to cell apoptosis.
Neurons contain intracellular deposits of ubiquitinated fibrillary proteins, a hallmark feature of various human neurodegenerative diseases such as Alzheimer's and Parkinson's. Engelender and colleagues identified an interaction amongst ubiquitin ligase E3 SIAH-1, SIAH-2, and synphilin-1 within Lewy bodies. The synphilin-1/SIAH complex, resistant to hydrolysis, results in the formation of a considerable quantity of ubiquitinated cellular inclusions. Analogous to the situation with synphilin-1, inside intact cells, SIAH-2 acts up on α-synuclein causing monoubiquitination, a state in which the protein is resistant to specific proteolytic enzyme. If the ubiquitin-proteasome system is unable to hydrolyze the protein, this leads to intracellular inclusion formation.
Layfield and others offered a novel interpretation for ubiquitinated inclusions at the molecular level: the presence of ubiquitinated proteins within inclusions does not necessarily indicate disorders arising from ubiquitin-mediated protein degradation. Instead, it could potentially be a secondary, protective cellular response. Ubiquitination is likely a subsequent process. Under such a hypothetical model, other factors might more readily promote inclusion formation.
Ubiquitination is a significant form of histone modification. Histones H2A and H2B are commonly ubiquitinated, and enzymes tasked with the ubiquitination of histone H2B have been discovered. Furthermore, a link between ubiquitination of H2B histones and histone methylation has been detected. Wang and colleagues purified a type of E3, hPRC1L (human polycomb repressive complex 1-like), and validated its function, finding that hPRC1L specifically targets histone H2A, ubiquitinating lysine 119 on the nucleosomal histones of H2A. Moreover, lysine 123 at the C-terminal of S. cerevisiae H2B is a substrate for E3 Rad6, the modification of which is crucial for both meiosis and mitosis. TafII 250, a component of the TBP-related complex, has been shown to invoke ubiquitin modification on H1, potentially implicating transcription processes.
Ubiquitination of activators plays a crucial role in transcription activation. In 2001, Salghetti et al. proposed that ubiquitination could serve as a bifunctional signal for both activation and restructuring of activators, regulating the functionality of Transactivation Domains (TAD). The E3 ubiquitin ligase Met30 mediates the ubiquitination of transcription activator VP16 TAD.
In the study of p53, it was discovered that HAUSP (Herpes Virus-Associated Ubiquitin Specific Protease) can deubiquitinate p53 both in vivo and in vitro, and stabilize p53 even in the presence of Mdm2. Both in vivo and in vitro, COP1 can bind specifically to p53 as a ubiquitin-conjugating enzyme, and then degrades it through the ubiquitin-proteasome system, preventing p53-mediated transcription and apoptosis.
References
- Hilt W, Wolf D H. The ubiquitin-proteasome system: past, present and future. Cellular and Molecular Life Sciences, 2004, 61: 1545
- Fang S, Weissman A M. A field guide to ubiquitylation. Cellular and Molecular Life Sciences, 2004, 61: 1546~1561
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem, 1998, 67: 425~479
- Pickart C M. Mechanisms underlying ubiquitination. Annu Rev Biochem, 2001, 70: 503~533
- Weissman A M. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol, 2001, 2: 169~178
- Wilkinson K D. Ubiquitination and deubiquitination: targeting of proteins for degradation by the proteasome. Semin Cell Dev Biol, 2000, 11: 141~148
- Bence N F, Sampat R M, Kopito R R. Impairment of the ubiquitin-proteasome system by protein aggregation. Science, 2001, (292): 1552~1555
- Spillantini M G, Schmidt M L, Lee V M, et al. Α-synuclein in Lewy bodies. Nature, 1997, 388: 839~840
- Engelender S, Kaminsky Z, Guo X, et al. Synphilin-1 associates with α-synuclein and promotes the formation of cytosolic inclusions. Nat Genet, 1999, 22: 110~114
- Layfield R, Cavey J R, Lowe J. Role of ubiquitin-mediated proteolysis in the pathogenesis of neurodegenerative disorders. Ageing Research Reviews, 2003, 2: 343~356
- Robzyk K, Recht J, OsleyM A. Rad6-dependent ubiquitination of histone H2B in yeast. Science, 2000, 287: 501~504
- Hwang W W, Venkatasubrahmanyam S, Ianculescu A G, et al. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol Cell, 2003, 11: 261~266
- Wood A, Krogan N J, Dover J, et al. An E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a pro-
- Sun Z W, Allis C D. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature, 2002, 418: 104~108
- Wang H B, Wang L J, Erdjument-Bromage H, et al. Role of histone H2A ubiquitination in polycomb silencing. Nature, 2004, 431: 873~878
- Pham A D, Sauer F. Ubiquitin-activating/conjugating activity of TAFII250, a mediator of activation of gene expression in drosophila. Science, 2000, 289: 2357~2360
- Salghetti S E, Caudy A A, Chenoweth J G, et al. Regulation of transcriptional activation domain function ubiquitin. Science, 2001, 293: 1651~1653
- Li M, Chen D, Shiloh A, et al. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature, 2002, 416: 648~653
- Dornan D, Wertz I, Shimizu H, et al. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature, 2004, 429(6987): 86~92
Our products and services are for research use only.