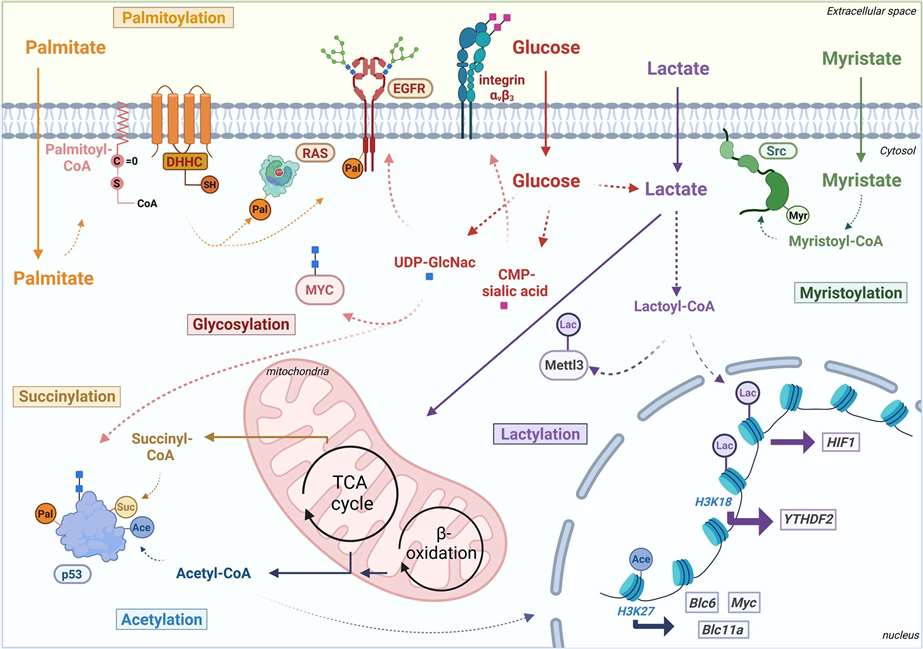
Integrative multi-omics approaches have emerged as a frontier in the field of omics, providing significant advantages by enabling comprehensive analysis of biological phenomena across different layers. This holistic perspective allows for the discovery of intricate information that is often inaccessible through single-omics studies. Recent integrated analyses of transcriptomic and proteomic data have revealed a limited correlation between the two, underscoring the fact that transcription, as an upstream regulator of protein expression, influences only a subset of protein levels. This observation highlights the necessity for further exploration of complex post-transcriptional regulatory mechanisms, particularly those involved in protein degradation pathways.
The Ubiquitin-Proteasome Degradation System
The ubiquitin-proteasome system (UPS) is the primary pathway for protein degradation in eukaryotic organisms, underscoring its critical importance. Recent studies have demonstrated that non-degradative regulatory roles mediated by the UPS are also crucial in biological signal transduction, especially within immune signaling pathways. Despite its significance, systematic omics studies focusing on both degradative and non-degradative ubiquitylation modifications remain scarce. This paucity of research further accentuates the novelty and expansive potential of the field of ubiquitylation proteomics.
Case Study
Integrative proteomics reveals an increase in non-degradative ubiquitylation in activated CD4+T cells
Journal: Nature Immunology
Impact Factor: 23.530
Integrative analysis of ubiquitination, proteomics, and transcriptomics, focusing on "ubiquitylation," has been employed to predict ubiquitin-mediated degradative and non-degradative modifications in T cells. This study elucidates the crucial role of ubiquitylation in T cell activation.
Introduction
CD4+ T cells play a pivotal role in adaptive immune responses. In the canonical T cell receptor (TCR) signaling pathway, ubiquitylation facilitates the degradation of key proteins via the proteasome and participates in signal transduction through non-degradative modifications. However, it remains unclear whether degradative or non-degradative ubiquitylation predominates in T cell activation. Thus, a comprehensive analysis of ubiquitylation types and functions in T cells is of significant importance.
Research Subjects
- Resting CD4+ T cells
- Activated CD4+ T cells (stimulated with CD3 and CD28)
Research Pathway
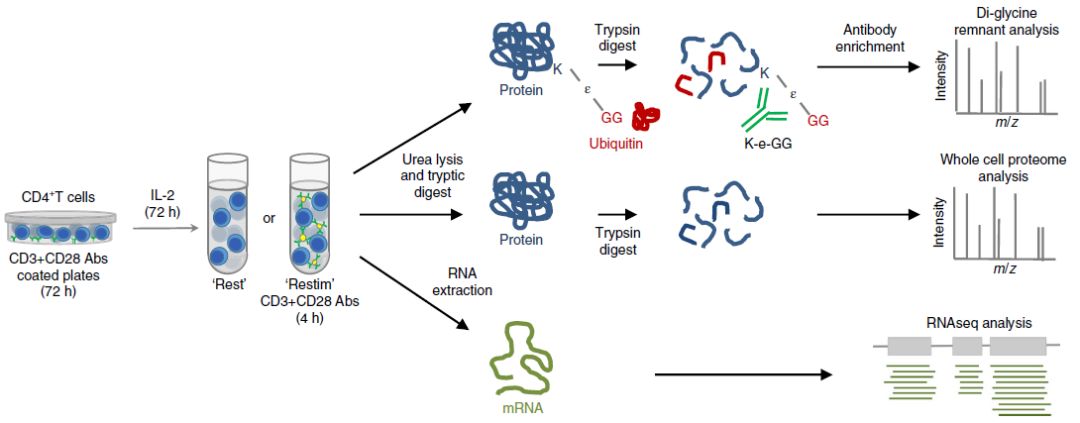 Integrative Proteomics and Transcriptomics Analysis of Ubiquitylation in CD4+ T Cells
Integrative Proteomics and Transcriptomics Analysis of Ubiquitylation in CD4+ T Cells
Results and Discussion
1. Proteomics Detection and Analysis
Utilizing comprehensive proteomics techniques, the study analyzed CD4+ T cells, identifying over 5,500 proteins (Figure 1). Among these, 4,556 proteins (71% of the total identified proteins) were consistently detected across three experimental groups. Upon activation of CD4+ T cells, 399 proteins exhibited upregulated expression, while 342 proteins displayed downregulated expression. Pathway analysis revealed significant responses in TCR and CD28 immune-related signaling pathways following T cell activation.
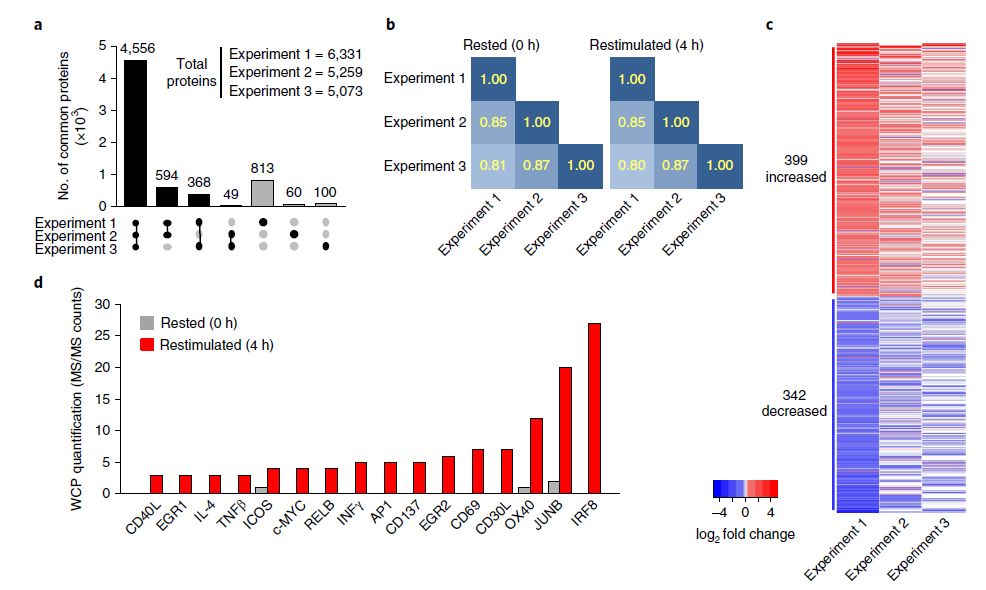 Figure 1: Proteomic analysis results of CD4+ T cells following activation.
Figure 1: Proteomic analysis results of CD4+ T cells following activation.
2. Integrative Analysis of Transcriptomics and Proteomics
Further transcriptomic analysis, integrated with proteomic data (Figure 2), revealed that 5,475 proteins (97% of the identified proteome) could be correlated with corresponding genes. Correlation analysis between transcriptomics and proteomics data indicated a poor correlation between mRNA and protein levels, consistent with previous reports.
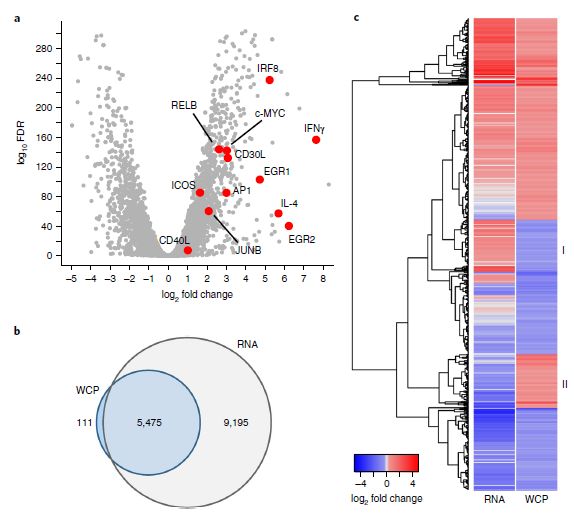 Figure 2: Comparative analysis of transcriptomic and proteomic alterations in CD4+ T cells post-activation.
Figure 2: Comparative analysis of transcriptomic and proteomic alterations in CD4+ T cells post-activation.
3. Integrative Analysis of Ubiquitinome and Proteome
Subsequently, ubiquitination profiling was performed to analyze changes in the ubiquitinome of CD4+ T cells. The integration of ubiquitinome and proteome data identified 1,080 proteins quantified in both datasets (Figure 3). Differential analysis revealed increased ubiquitination of 221 proteins upon T cell activation, with 43 proteins uniquely ubiquitinated in activated CD4+ T cells. Conversely, 217 proteins exhibited decreased ubiquitination, with 121 proteins uniquely ubiquitinated in resting CD4+ T cells.
Correlation Between Ubiquitination and Protein Abundance
Correlation analysis indicated that proteins such as CD3ε, ZAP-70, and LAT were downregulated in the proteome but upregulated in the ubiquitinome, suggesting potential degradation. However, several proteins showed concordant trends in both ubiquitination and abundance changes. Overall, the ubiquitinome did not display a strong negative correlation with total protein changes, indicating that TCR-induced ubiquitination modifications may not predominantly lead to protein degradation.
Our hot services
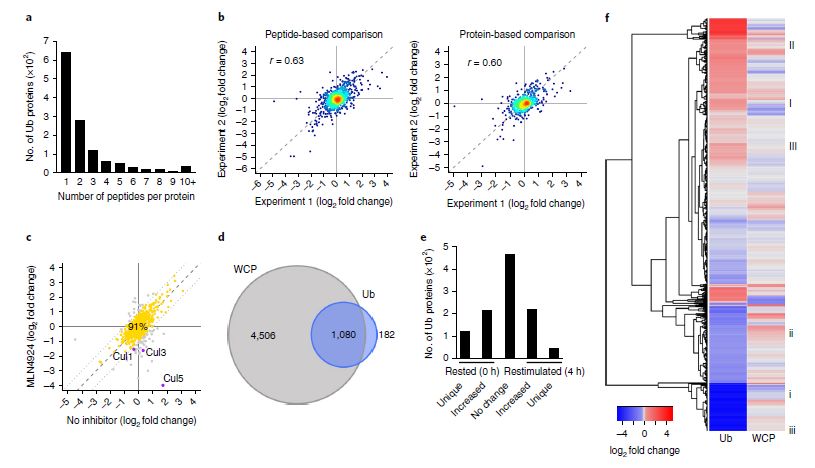 Figure 3: Differential ubiquitinome and proteome changes in CD4+ T cells following activation.
Figure 3: Differential ubiquitinome and proteome changes in CD4+ T cells following activation.
4. Integrative Analysis of Ubiquitinome, Proteome, and Transcriptome in TCR Signaling Pathway
Selection and Analysis of Signaling Proteins
The study focused on 155 signal proteins within the TCR signaling pathway. The integration of transcriptomic, proteomic, and ubiquitinome data identified significant regulatory changes (Figure 4).
Ubiquitination Dynamics and Protein Stability
Among the 31 proteins undergoing ubiquitination modifications, 12 proteins exhibited increased ubiquitination in activated CD4+ T cells. Integrative analysis with transcriptomic and proteomic data predicted degradation-associated ubiquitination for NFKB1, UBE2N, ZAP-70, and LAT, characterized by elevated transcription and ubiquitination levels, alongside decreased or stable protein abundance.
Conversely, proteins such as CD3ε, CD3γ, CD3ζ, PKCθ, CDC42, RAC1, RHOA, and GRAP demonstrated concordant changes in RNA and protein expression levels, suggesting that their ubiquitination did not lead to protein degradation.
Validation of Ubiquitination Predictions
Validation experiments confirmed the predicted ubiquitination outcomes for ZAP-70 and PKCθ. ZAP-70 was found to undergo degradation-associated ubiquitination, while PKCθ displayed non-degradation ubiquitination, corroborating the integrative analysis results.
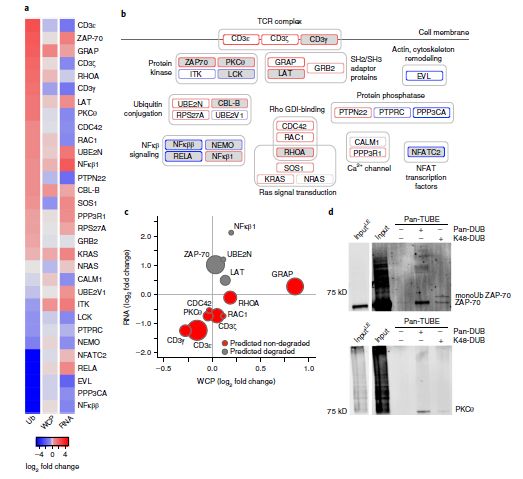 Figure 4: Differential changes in the ubiquitinome and proteome of CD4+ T cells following activation.
Figure 4: Differential changes in the ubiquitinome and proteome of CD4+ T cells following activation.
5. Role of Non-Degradative Ubiquitination in T Cell Activation
Quantitative Analysis of Ubiquitination Modifications
To assess the proportion and function of non-degradative versus degradative ubiquitination modifications during T cell activation (Figure 5), a predictive framework for the TCR signaling pathway was employed. The study quantified all detected proteins to predict their ubiquitination status.
Increased Non-Degradative Ubiquitination
Upon activation of CD4+ T cells, there was a notable increase in non-degradative ubiquitination modifications. Specifically, the abundance of K29, K33, and K63 non-degradative ubiquitination peptides increased significantly in activated CD4+ T cells. In contrast, the abundance of K48 and K11 ubiquitination peptides, typically associated with proteasomal degradation, remained unchanged.
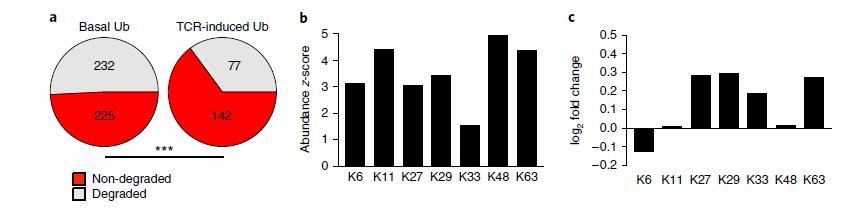 Figure 5: Differential changes in the ubiquitinome and proteome of CD4+ T cells following activation.
Figure 5: Differential changes in the ubiquitinome and proteome of CD4+ T cells following activation.
Conclusion
This study utilized an integrative approach combining transcriptomics, proteomics, and ubiquitin-proteomics to develop a predictive method for identifying types of ubiquitination modifications. The findings elucidate the pivotal role of non-degradative ubiquitination in T cell activation following T cell receptor (TCR) stimulation, providing a robust theoretical foundation for further exploration of T cell ubiquitination functions.
Broader Implications and Applications
The predictive framework developed in this study demonstrates extensive applicability, extending its relevance beyond T cell research to encompass other biological systems. This framework holds substantial promise for deepening our understanding of ubiquitination across diverse contexts, including disease mechanisms and therapeutic strategies. By offering a detailed perspective on ubiquitination dynamics, this research sets the stage for future investigations aimed at translating these insights into clinical applications.
Reference
- Dybas, J.M., O'Leary, C.E., Ding, H. et al. Integrative proteomics reveals an increase in non-degradative ubiquitylation in activated CD4+ T cells. Nat Immunol 20, 747–755 (2019).
Our products and services are for research use only.