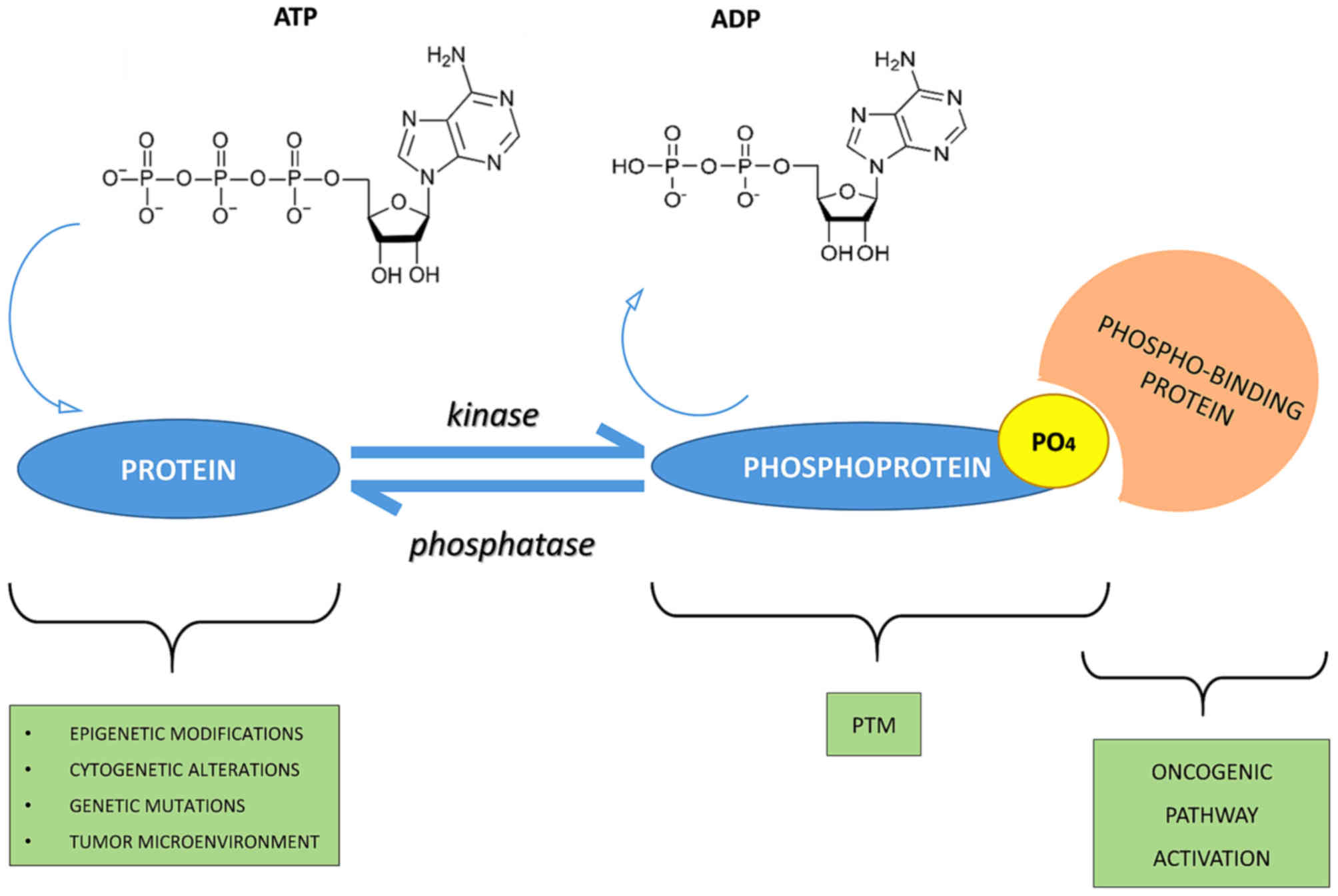
Author: Creative Proteomics scientific team — senior proteomics scientists and bioinformaticians (Creative Proteomics). Specialties: Orbitrap DIA/TMT phosphoproteomics with IMAC/TiO2 enrichment. See our service overview: Creative Proteomics phosphoproteomics services.
The PI3K–AKT–mTOR axis orchestrates cell survival, growth, and metabolism through a tightly regulated protein phosphorylation cascade. For translational oncology, single-node readouts (e.g., Western blots of p-AKT) are no longer sufficient to validate kinase inhibitor efficacy. Instead, site-specific, quantitative mapping of akt protein phosphorylation across the activation hub (Thr308 and Ser473) and downstream effectors (PRAS40 Thr246, GSK3β Ser9, TSC2 Thr1462) provides a mechanistic, auditable picture of target engagement and pathway rewiring across tumor cohorts.
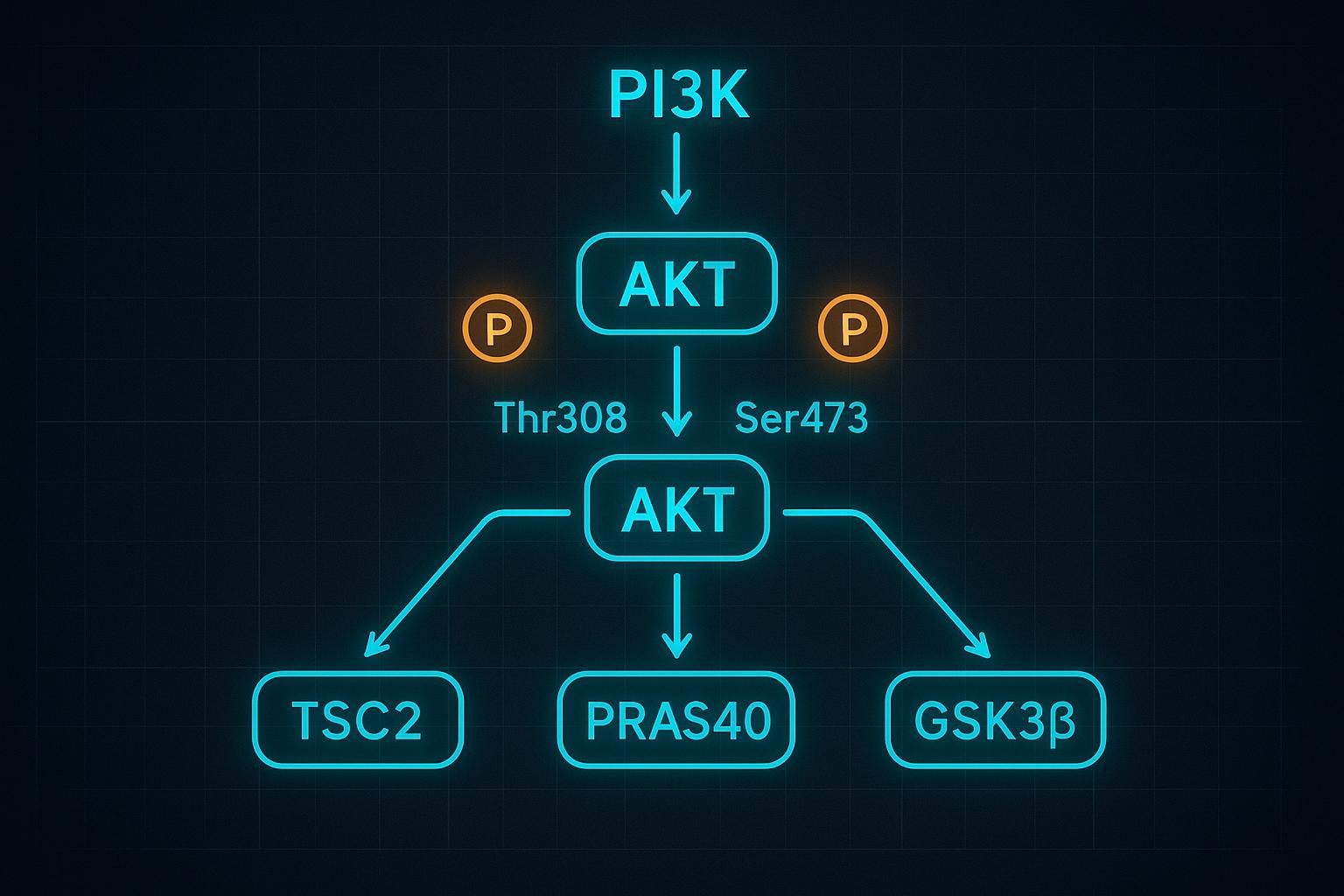
Deciphering the PI3K/AKT/mTOR Axis: A Multi-Node Protein Phosphorylation Cascade
The Initiation of the AKT Signaling Network
Membrane recruitment via PI3K-generated PIP3 brings AKT and its upstream kinases into proximity. PDK1 phosphorylates AKT at Thr308 within the activation loop, while mTORC2 phosphorylates Ser473 in the hydrophobic motif, together enabling full catalytic competence and substrate selectivity. Dephosphorylation is executed by PP2A (Thr308) and PHLPP (Ser473), closing the cycle. A 2020 open-access synthesis by Balasuriya and colleagues details the dual-site activation mechanics and substrate-tuning role of Ser473—see the mechanistic overview in Balasuriya et al., 2020 (PNAS/PMC). For broader cancer signaling context, Peng et al., 2022 reviews PI3K/AKT/mTOR regulation and oncogenic implications.
Molecular Drivers of the Pathological Protein Phosphorylation Cascade
Oncogenic PI3K alterations or PTEN loss elevate PIP3 levels, increasing AKT recruitment and phosphorylation. The result is persistent akt protein phosphorylation and amplified signaling through mTORC1 and other branches governing translation, apoptosis resistance, and cell-cycle progression. Compartmentalization at endosomes and lysosomes further modulates signaling kinetics and substrate access (e.g., Chu et al., eLife, 2020; Kim et al., 2021 (PMC)).
Dual Phosphorylation Requirements: Thr308 and Ser473 Activation
- Thr308 (PDK1): activation loop phosphorylation that initiates catalytic activity.
- Ser473 (mTORC2): hydrophobic motif phosphorylation that fine-tunes substrate specificity and maximal activity; also influences PDK1 efficiency.
- Phosphatases: PP2A (Thr308) and PHLPP (Ser473) reset the kinase.
Downstream Effectors: TSC2, GSK3β, and PRAS40 Feedback Loops
- TSC2 Thr1462: AKT phosphorylation inhibits the TSC complex, activating Rheb→mTORC1 and boosting translation initiation (Jia et al., 2019 (PMC)).
- GSK3β Ser9: phosphorylation inactivates GSK3β, affecting glycogen metabolism and numerous transcriptional programs (Palma et al., 2023 (PMC)).
- PRAS40 Thr246: phosphorylation relieves its inhibitory brake on mTORC1, integrating with mTORC1-dependent events (Mi et al., 2015 Oncotarget; Chen et al., 2021 (PMC)).
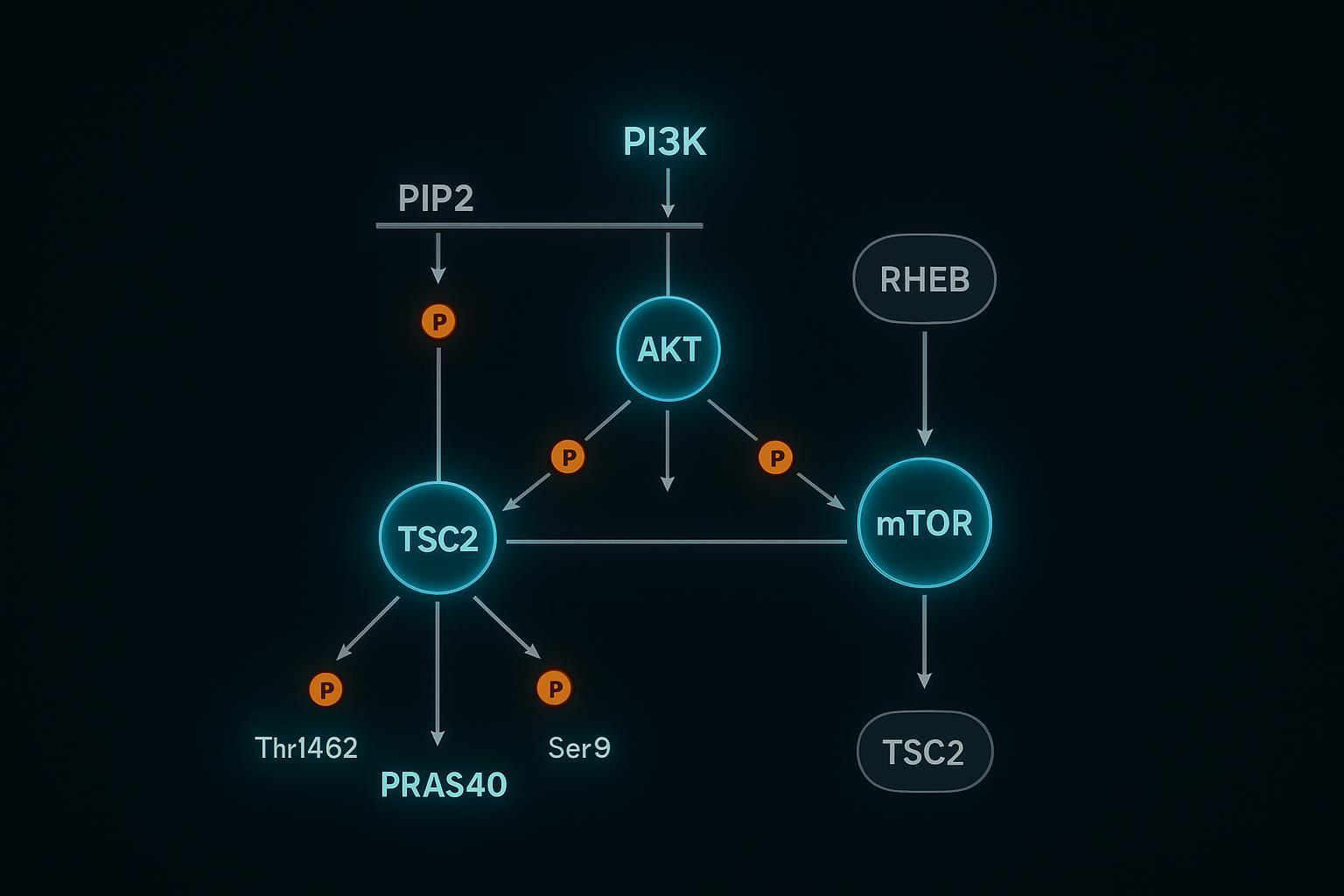 Figure 1: The intricate protein phosphorylation cascade of the AKT pathway, highlighting critical activation nodes for oncology research.
Figure 1: The intricate protein phosphorylation cascade of the AKT pathway, highlighting critical activation nodes for oncology research.
Overcoming Complexity: High-Sensitivity Orbitrap MS Detection for Low-Abundance AKT Sites
The Limitation of Traditional Immunoassays in Pharma R&D
Antibody-based assays such as Western blots struggle with cross-reactivity, narrow dynamic ranges, and poor detectability when site occupancy drops below a few percent. These constraints limit confident quantitation of low-abundance phosphorylation events and multiplexed readouts across a pathway. Method comparisons consistently show that LC–MS/MS offers antibody-independent, site-specific quantitation with wider linear ranges and better multiplexing for pathway-level decisions. For an accessible overview of Western blot pitfalls (linearity, saturation, stripping artifacts), see Bass et al., 2016 (PMC).
Advanced LC-MS/MS Strategies for Deep Phosphoproteome Coverage
Orbitrap-based DIA provides broad coverage and high reproducibility suitable for cohort-scale pathway mapping, while targeted PRM delivers the highest precision for specific AKT nodes. Gas-phase fractionation DIA can match targeted-mode reproducibility for many peptides while retaining discovery breadth—see the methodological performance summary in Searle et al., 2023 (PMC). For PRM in complex matrices, parameters and best practices are outlined in Dakup et al., 2023 (PMC).
For instrument setup reference (windowing schemes, Orbitrap resolution settings, and cycle timing), see Creative Proteomics’ Orbitrap MSX-DIA parameters and capabilities overview.
Immunoaffinity Enrichment Combined with Orbitrap Mass Spectrometry
- Enrichment toolkit: IMAC and TiO2 are complementary for global phosphopeptide capture; motif-directed immunoaffinity precipitation (IAP) can boost recovery of specific phospho-motifs. Sequential or orthogonal enrichment often yields deeper coverage (e.g., Possemato et al., 2017 (PMC); Kitata et al., 2021 (PMC)).
- Fractionation: SAX/SCX/HILIC prior to LC–MS/MS can improve depth when sample permits.
- Practical note: preserve site localization confidence by optimizing acid strength and minimizing non-specific carryover.
Quantitative Precision: Achieving Low LOD/LOQ in Complex Matrixes
- Pair PRM on a quadrupole–Orbitrap with SIS peptides for Thr308, Ser473, PRAS40 Thr246, GSK3β Ser9, and TSC2 Thr1462.
- Typical targets in clinical matrices (plasma/CSF/tissue digests) aim for low fmol to amol LOD/LOQ, with linearity spanning 3–4 orders of magnitude when matrix-matched calibrators are used.
- Optimize resolution (≥30k–60k), isolation width (1–2 m/z), AGC (1e5–1e6), and max injection time (30–100 ms) to maximize sensitivity without compromising selectivity.
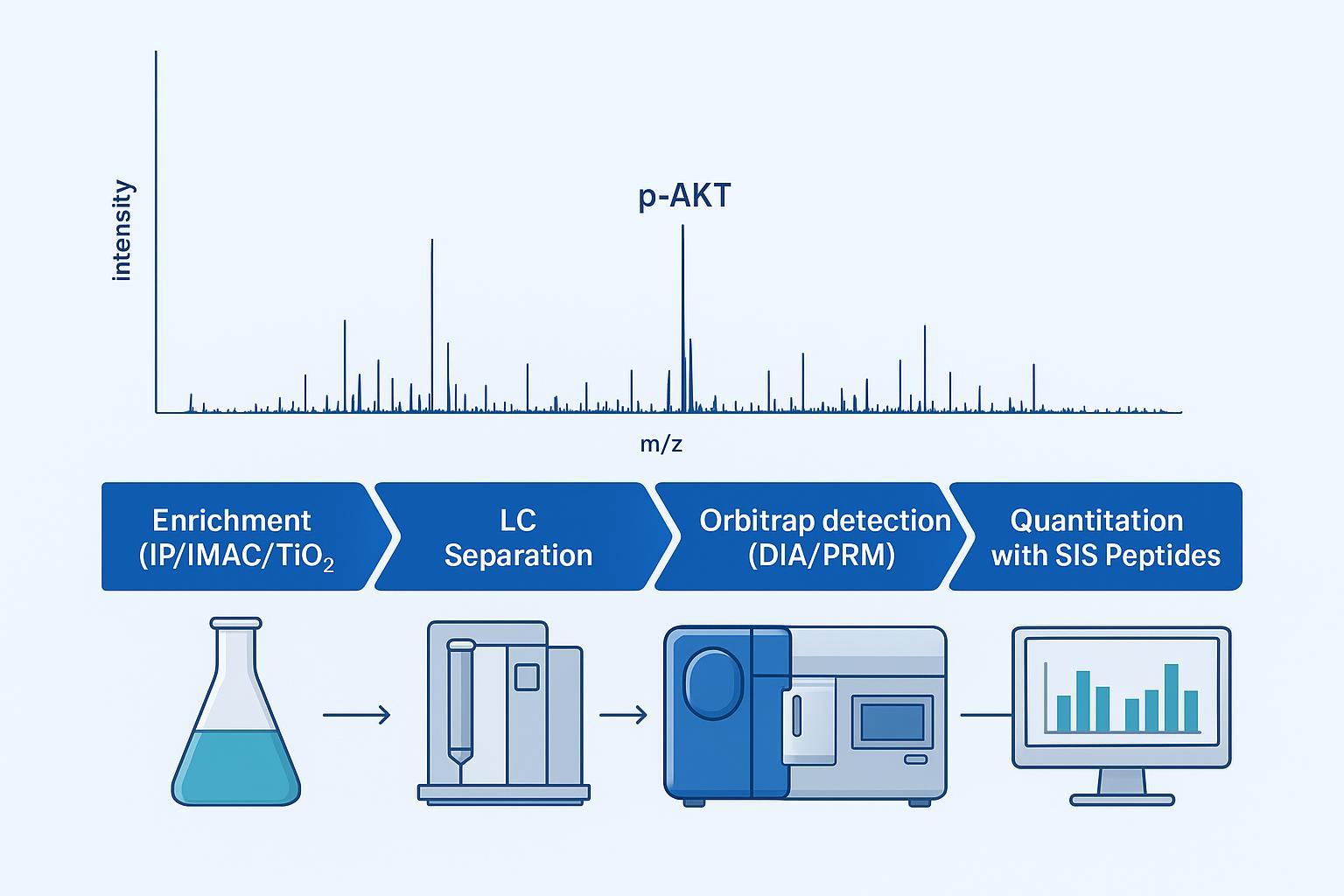 Figure 2: Utilizing high-resolution Orbitrap MS for the quantitative protein phosphorylation analysis of AKT isoforms in clinical samples.
Figure 2: Utilizing high-resolution Orbitrap MS for the quantitative protein phosphorylation analysis of AKT isoforms in clinical samples.
Methodological Rigor: Validating Kinase Inhibitor Potency with Site-Specific Quantitation
Establishing Robust QC Metrics for Clinical-Grade Proteomics
For oncology PDX or biopsy samples (frozen tissue or FFPE; input ≥10–20 mg), define auditable QC up front:
- Reproducibility: Intra-batch CV ≤10%; Inter-batch CV ≤15%.
- Sensitivity: LOD/LOQ at low fmol–amol for targeted AKT phosphopeptides.
- Linearity: 3–4 orders of magnitude in matrix-matched calibration.
- SIS calibration: absolute quantitation deviation ≤ ±20% for standards and QCs.
- Site localization: adopt stringent thresholds (e.g., high-confidence Ascore or PTMProphet probabilities) and control global false localization rates (see localization guidance in [Slavov et al., 2022/2024 methodological discussions] and PTMProphet best-practices, 2022).
Reproducibility Standards: Intra-batch and Inter-batch CV Targets
- Design QC sets: pooled matrix controls, SIS-spiked QCs at low/med/high levels, and replicate injections to estimate precision.
- Acceptance bands: Per-site intra-batch CV ≤10% (target) and inter-batch ≤15% (target) after normalization. Investigate outliers with retention-time drift, interference, or localization ambiguity. Cohort proteomics studies demonstrate achievable CV bands following rigorous normalization (Gerritsen et al., 2021 (PMC)).
Dynamic Range and Linearity in Site-Specific Phospho-Quantification
- Build 6–8 point matrix-matched calibration curves per SIS/peptide pair; evaluate R², back-calculated accuracies, and residual plots.
- Confirm linearity across 3–4 orders of magnitude. Use weighted regression (1/x or 1/x²) to stabilize low-end fit.
- Determine LOD/LOQ using blank variance (e.g., 3.3σ/s and 10σ/s rules) or ICH-based criteria; verify at the method level with independent QCs.
Stable Isotope Labeling and Synthetic Phosphopeptide Calibration Strategies
- SIS peptides: design sequence/charge-matched labeled analogs for AKT Thr308/Ser473 tryptic peptides and for PRAS40 Thr246, GSK3β Ser9, TSC2 Thr1462 sites.
- Co-elution and transition selection: confirm co-elution and match fragment ion ratios; monitor interferences and adjust isolation or transitions as needed.
- Traceability: document lot numbers, purity, and concentration assignments; keep calibration records to support audit trails.
CRO Strategic Insights: Accelerating NDA/IND Filings via Transparent Data Governance
Milestone Management in Large-Scale Oncology Cohorts
- Pre-analytical: finalize tissue handling (macrodissection, fixation/deparaffinization for FFPE), estimate peptide yields from 10–20 mg inputs, and lock enrichment strategy (IMAC/TiO2 ± IAP).
- Analytical: define acquisition plans (DIA for profiling vs PRM for absolute quant), retention-time alignment, and localization scoring thresholds.
- Post-analytical: deliver raw files, search outputs, site tables with localization probabilities, calibration reports, QC summaries, and audit logs.
Integrating Multi-PTM Evidence: AKT Phosphorylation and Ubiquitylation Interplay
- Crosstalk rationale: AKT activity and substrate fate can be modulated by ubiquitylation; conversely, phosphorylation can gate ubiquitin ligase interactions. Including both PTMs refines mechanism-of-action claims. For general methods on ubiquitin remnant enrichment, see [Ub-remnant (K-ε-GG) workflows, review examples].
- Practical approach: use phospho-centric DIA/PRM plus affinity enrichment for ubiquitinated peptides to build convergent evidence.
Regulatory Compliance and Data Integrity for Pharma Partners
- Adopt ALCOA+ data integrity principles with validated pipelines and complete audit trails. Align electronic records and signatures with the FDA Data Integrity guidance (2018) and FDA Part 11 scope/application; for clinical systems, refer to the EMA computerized systems guideline (2023).
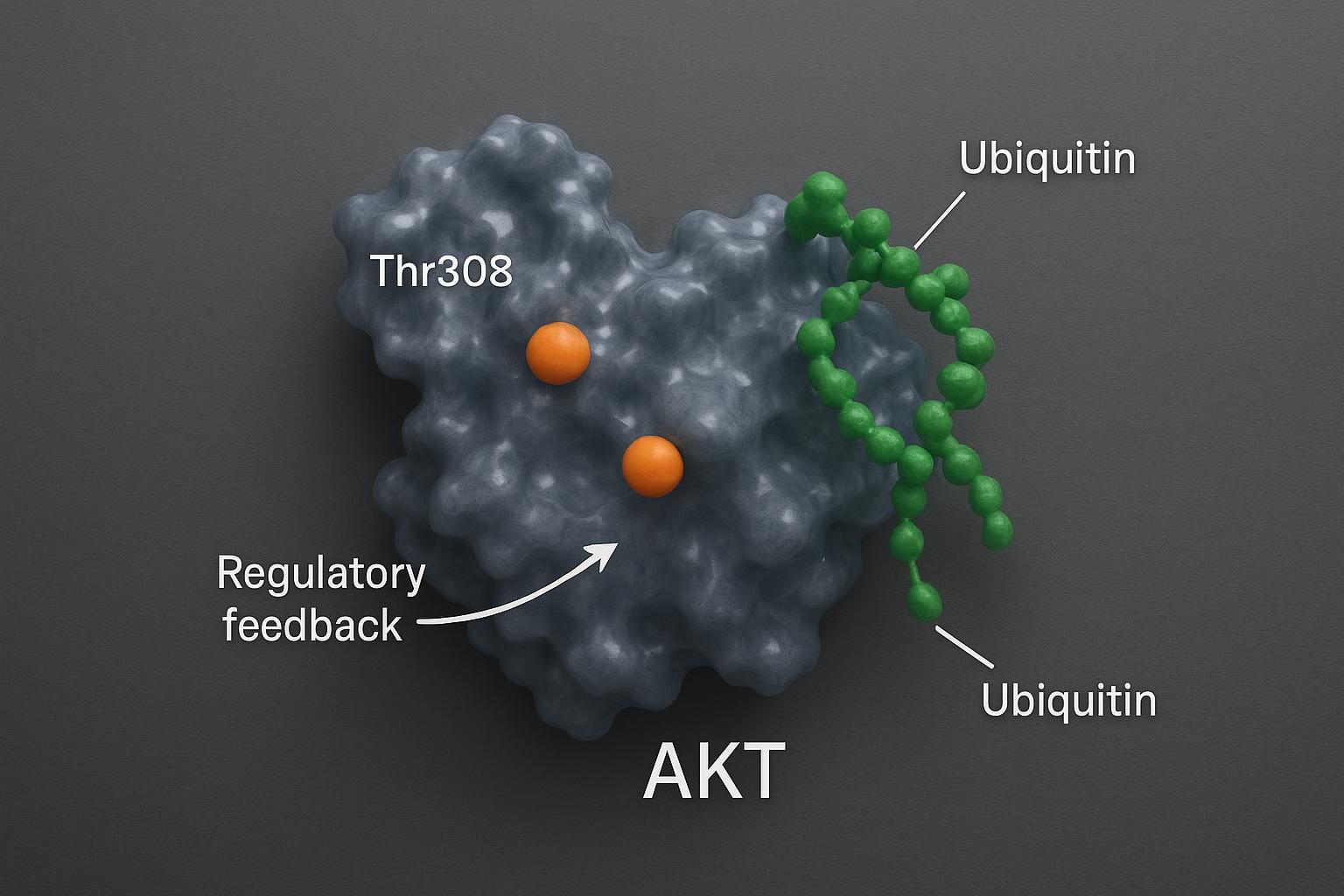 Figure 3: Integrating evidence of akt protein phosphorylation and ubiquitylation crosstalk for comprehensive drug efficacy profiling.
Figure 3: Integrating evidence of akt protein phosphorylation and ubiquitylation crosstalk for comprehensive drug efficacy profiling.
Future Directions: Mapping the Global Kinome for Next-Gen AKT Inhibitors
Leveraging Global Protein Phosphorylation Databases for Target Validation
Public phosphoproteomics resources and spectral libraries can accelerate validation of AKT substrates and context-dependent signaling. Kinome-wide DIA maps coupled with targeted PRM verification sharpen hypotheses about resistance and adaptive rewiring.
From CSF Markers to Companion Diagnostics
As sensitivity improves, biofluid phosphoproteomics (e.g., CSF) may support exploratory markers feeding into companion diagnostic concepts. Any clinical trajectory demands stringent site-specific quantitation, orthogonal verification, and prospective validation under governance suitable for regulated use.
For teams seeking outsourced, Orbitrap-based PTM workflows with site-level quantitation and governance aligned to the QC thresholds outlined here, a service provider such as Creative Proteomics can be considered. See the overview of PTM analysis fundamentals for background context: Comprehensive PTM analysis.
References and further reading:
- Dual activation and substrate tuning: Balasuriya et al., 2020 (PMC); broader PI3K/AKT/mTOR context: Peng et al., 2022 (PMC).
- Compartmentalization and signaling localization: Chu et al., 2020, eLife; Kim et al., 2021 (PMC).
- Immunoassay pitfalls: Bass et al., 2016 (PMC); tutorial: Azure Biosystems, 2021.
- Orbitrap DIA/PRM methods: Searle et al., 2023 (PMC); Dakup et al., 2023 (PMC).
- Enrichment strategies: Possemato et al., 2017 (PMC); Kitata et al., 2021 (PMC).
- Localization confidence and data integrity: PTMProphet best-practices, 2022 (PMC); FDA Data Integrity (2018); EMA computerized systems (2023).
Our products and services are for research use only.



.jpg)