While cancer research has long centred on genetic mutations, scientists are increasingly recognising the importance of epigenetic changes—alterations that affect how genes are turned on or off without changing the DNA itself. One key type of epigenetic change involves histone modifications, which are chemical tags added to the proteins that package DNA. These tags help control which genes are active or silent. Unlike permanent genetic mutations, histone modifications can be reversed, which makes them promising targets for new cancer treatments. By studying changes in these histone proteins, researchers can uncover valuable clues for earlier diagnosis, identify potential biomarkers, and design more precise therapies.
Explore how we support cancer epigenetics research with our Histone PTM Analysis service.
What Are Histone Modifications?
Histone modifications are chemical alterations to histone proteins—primarily H2A, H2B, H3, and H4—that regulate chromatin structure and gene expression. These modifications occur predominantly on the N-terminal tails of histones and are catalysed by specific enzymes. They include:
- Acetylation (e.g., H3K27ac): Neutralises the positive charge on lysines, loosens chromatin structure, and facilitates transcription.
- Methylation (e.g., H3K4me3, H3K27me3): Can either activate or repress genes, depending on the residue and methylation state (mono-, di-, or tri-methylation).
- Phosphorylation (e.g., H3S10ph): Associated with chromosome condensation, DNA repair, and cell cycle progression.
- Ubiquitination (e.g., H2BK120ub): Plays roles in transcription elongation and DNA damage response.
- SUMOylation: Generally represses transcription by promoting chromatin compaction.
Select Service
 Figure 1. Histone modifications in cancer (Xu X, et al., 2023).
Figure 1. Histone modifications in cancer (Xu X, et al., 2023).
Histone Modifiers and Their Dysregulation in Cancer
Histone modifications are dynamically regulated by three classes of enzymes:
- Writers (e.g., histone acetyltransferases [HATs], histone methyltransferases [HMTs])
- Erasers (e.g., histone deacetylases [HDACs], histone demethylases [KDMs])
- Readers (e.g., bromodomain and chromodomain proteins)
In cancer, mutations, overexpression, or loss-of-function in these enzymes lead to widespread epigenetic reprogramming. For example:
- EZH2, a catalytic subunit of the Polycomb Repressive Complex 2 (PRC2), adds H3K27me3 to silence tumor suppressors. Gain-of-function mutations in EZH2 are found in lymphomas and drive oncogenic silencing.
- KDM6A/UTX, a demethylase that removes H3K27me3, is frequently inactivated in bladder and pancreatic cancers, leading to aberrant repression.
- CREBBP and EP300, key HATs, are mutated in hematologic malignancies, resulting in reduced histone acetylation and impaired transcriptional activation of differentiation genes.
- HDAC overexpression is common in solid tumours, leading to global hypoacetylation and transcriptional repression of genes involved in apoptosis and cell cycle arrest.
Related Article
The Most Comprehensive Overview of Histone Modification
Histone PTMs as Cancer Biomarkers
PTMs are increasingly recognised as valuable epigenetic biomarkers for cancer diagnosis, prognosis, and therapy selection. Unlike genetic mutations, histone PTMs reflect dynamic changes in gene regulation and tumor microenvironment, making them sensitive indicators of disease state and treatment response.
Specific histone marks are associated with distinct cancer phenotypes:
- H3K27me3 loss is frequently observed in malignant peripheral nerve sheath tumours (MPNSTs) and correlates with aggressive tumor behavior (Lee W, et al., 2014).
- Global H4K20me3 reduction is linked to genomic instability and poor prognosis in lung, breast, and colon cancers (Fraga MF, et al., 2005).
- H3K9me2 and H3K9ac levels differentiate between low- and high-grade gliomas, offering potential for tumor grading (Muller T, et al., 2012).
Quantitative profiling of these PTMs can aid in:
- Early cancer detection, especially in liquid biopsies
- Monitoring therapeutic response to epigenetic drugs (e.g., HDAC or EZH2 inhibitors)
- Stratifying patients for targeted therapies or clinical trials
Crosstalk Between Histone Modifications and Other Epigenetic Mechanisms
Histone modifications interact dynamically with other epigenetic layers, forming a coordinated regulatory network that governs chromatin architecture and gene expression in cancer.
Histone Modifications and DNA Methylation
There is a strong interplay between histone methylation and DNA methylation. For instance, repressive histone marks such as H3K9me3 and H3K27me3 often co-localise with 5-methylcytosine (5mC) at silenced gene promoters. Histone methyltransferases like SUV39H1 and EZH2 recruit DNA methyltransferases (DNMTs), reinforcing transcriptional repression. This cooperative silencing is a hallmark of epigenetically inactivated tumor suppressor genes.
Histone Modifications and Non-Coding RNAs
Long non-coding RNAs (lncRNAs) serve as scaffolds for histone-modifying complexes. For example, HOTAIR interacts with PRC2, guiding it to specific genomic loci to deposit H3K27me3 and silence metastasis-suppressor genes in breast cancer. This lncRNA–histone interaction exemplifies how non-coding RNAs orchestrate localised histone modifications and chromatin remodeling.
Histone Modifications and Chromatin Remodelers
Histone marks modulate the recruitment of chromatin remodeling complexes, which reposition or eject nucleosomes. Acetylated histones, particularly H3K27ac, serve as binding platforms for bromodomain-containing proteins (e.g., BRD4), which in turn facilitate transcriptional elongation at oncogenic enhancers. Conversely, repressive marks like H3K9me3 promote heterochromatin formation via recruitment of HP1.
Technologies for Profiling Histone Modifications in Cancer Samples
Accurate profiling of histone modifications is essential for decoding cancer-specific epigenetic states. Several complementary technologies are used to identify, localise, and quantify these PTMs:
Mass Spectrometry (MS)-Based Proteomics
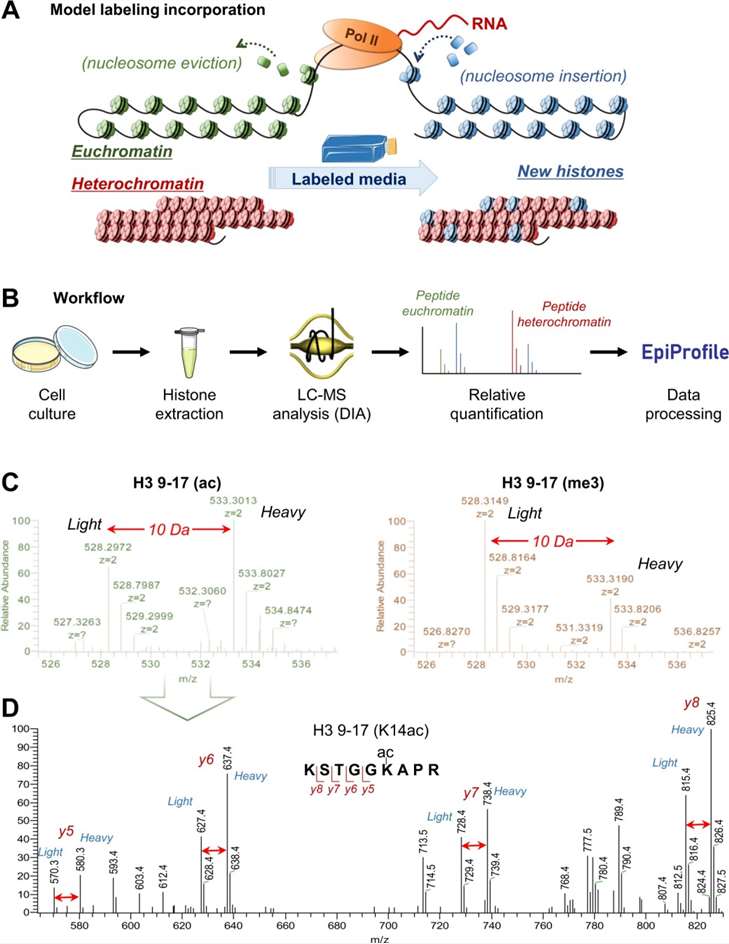
MS is the gold standard for comprehensive and quantitative analysis of histone PTMs. It enables high-resolution identification of multiple modifications on the same histone tail (e.g., combinatorial marks on H3 and H4), site-specific quantification, and comparison across cancer samples. When combined with stable isotope labeling (e.g., SILAC or TMT), MS allows accurate measurement of PTM abundance under different treatment or disease conditions.
Chromatin Immunoprecipitation Followed by Sequencing (ChIP-seq)
ChIP-seq maps the genomic locations of specific histone modifications by using modification-specific antibodies to enrich targeted nucleosomes. This method provides functional context by linking histone marks to active promoters, enhancers, or silenced chromatin regions. However, its accuracy depends heavily on antibody specificity and chromatin quality.
Cut&Run and Cut&Tag
These emerging techniques are antibody-guided and enzyme-assisted methods for mapping histone marks. They offer higher resolution, lower background noise, and require fewer cells than ChIP-seq, making them ideal for low-input cancer samples or rare cell populations.
Western Blotting and ELISA
While less comprehensive, these techniques offer quick, semi-quantitative readouts for selected histone modifications. They are often used for validation or for screening histone mark changes in treated cancer cells.
Select Service
Related Article
Case Studies: Histone PTM Profiling in Oncology Research
Acute Myeloid Leukemia (AML)
Case: Quantitative proteomic analysis of histone modifications in decitabine-sensitive and resistant leukemia cell lines
Background
Resistance to epigenetic drugs like decitabine (DAC) presents a significant challenge in the treatment of AML and myelodysplastic syndromes (MDS). DAC acts as a DNA methyltransferase inhibitor, yet its mechanism of action and the basis for drug resistance remain poorly defined. Given the crosstalk between DNA methylation and histone PTMs, this study investigates histone PTM patterns in DAC-sensitive and resistant leukemia cells to identify biomarkers and uncover epigenetic mechanisms involved in DAC response.
Purpose
- Identifying novel and differential histone PTMs,
- Uncovering PTM biomarkers predictive of DAC resistance,
- Investigating epigenetic differences between early- and late-stage AML cells.
Method
- Two leukemia cell lines (MDS-L and TF-1) were analysed in DAC-sensitive and DAC-resistant states.
- Histones were acid-extracted, chemically propionylated, and analysed via LC–MS/MS with SILAC-based internal standards for accurate quantification.
- A simplified histone enrichment method without HPLC or gel separation was developed.
- PTM analysis and quantification were performed using MaxQuant, targeting acetylation and methylation marks.
Results
- 61 individual histone marks and 60 PTM combinations were quantified.
- 15 novel PTMs were discovered, primarily involving mono-methylation on lysine residues across H1, H2A, H2B, and H4.
- Differential PTMs between DAC-sensitive and -resistant cells were identified:
In MDS-L cells: H3.3K36me3 and H4K8acK12acK16ac altered with DAC treatment.
In TF-1 cells: H3.1K27me1, H3.1K36me1, and H3.1K27me1K36me1 showed DAC-specific regulation.
- H3K27me2/3 and H3K36me2 were significantly more abundant in TF-1 (advanced AML) compared to MDS-L (early stage MDS), indicating potential biomarkers for leukemia progression.
- These PTMs implicate enzymes like EZH2 and histone acetyltransferases (HATs) as key players in epigenetic regulation during DAC treatment.
 Figure 2. Fold changes of histones and histone PTMs in two cell lines.
Figure 2. Fold changes of histones and histone PTMs in two cell lines.
Glioblastoma (GBM)
Case: Inhibition of Histone Deacetylation Potentiates the Evolution of Acquired Temozolomide Resistance Linked to MGMT Upregulation in Glioblastoma Xenografts
Background
Temozolomide (TMZ) is a frontline chemotherapeutic for GBM, but treatment is hampered by both primary and acquired resistance. MGMT repairs TMZ-induced DNA lesions; its high expression or lack of promoter methylation correlates with poorer patient outcomes. While MGMT promoter hypermethylation silences MGMT in many tumours, other epigenetic mechanisms—particularly histone modifications—may drive MGMT re-expression during resistance evolution.
Purpose
The study aimed to establish an in vivo GBM xenograft model of acquired TMZ resistance and to dissect mechanisms underlying TMZ resistance, with a focus on histone deacetylation and its inhibition by SAHA (suberoylanilide hydroxamic acid).
Method
- MGMT Analysis
Promoter methylation: MS-PCR and pyrosequencing of CpG sites.
Expression: qRT-PCR and Western blot for MGMT mRNA/protein.
- Chromatin Immunoprecipitation (ChIP)
Assessed H3K9-ac, H3K9-me2, and H3K27-me3 occupancy at the MGMT promoter.
Measured recruitment of Sp1, c-JUN, NF-κB, and p300.
- Functional Assays
In vitro cytotoxicity with and without the MGMT inhibitor O^6-benzylguanine (O^6-BG).
Neurosphere formation assays.
- Compared resistance evolution in GBM12 xenografts treated with TMZ alone versus TMZ + SAHA.
Results
- 3 of 5 resistant lines (GBM12TMZ, GBM14TMZ, GBM28TMZ) showed marked MGMT mRNA/protein re-expression despite unchanged promoter methylation. In these lines, O^6-BG restored TMZ sensitivity in vitro, confirming MGMT's causal role.
- Resistant tumours with MGMT re-expression displayed increased H3K9-ac and decreased H3K9-me2 at the MGMT promoter, with no change in H3K27-me3. Enhanced recruitment of Sp1, c-JUN, NF-κB, and p300 paralleled MGMT reactivation.
- TMZ + SAHA co-treatment extended tumor growth delay similarly to TMZ alone but favored MGMT-driven resistance: 5/8 recurrences in the combination group re-expressed MGMT versus none with TMZ monotherapy. Global H3K9-ac rose in all tissues with SAHA, but MGMT promoter acetylation—and consequent MGMT upregulation—was specific to resistant tumours from the combination arm.
 Figure 3. Evaluation of the chromatin marks within the MGMT promoter region comparing pair-matched parental and temozolomide-resistant GBM12, 14, and 22.
Figure 3. Evaluation of the chromatin marks within the MGMT promoter region comparing pair-matched parental and temozolomide-resistant GBM12, 14, and 22.
Relevant FAQ
How do histone modifications differ from genetic mutations in cancer?
Unlike permanent DNA mutations, histone modifications are reversible. This makes them attractive targets for therapeutic intervention and dynamic biomarkers for cancer diagnosis and prognosis.
What is the difference between DNA methylation and histone modifications?
DNA methylation directly modifies DNA and typically leads to gene silencing, while histone modifications alter chromatin structure and accessibility. Both regulate gene expression but via distinct molecular mechanisms.
How do I choose between ChIP-seq and mass spectrometry for histone PTM analysis?
Choose ChIP-seq when you need genome-wide location data of a specific PTM, and mass spectrometry when you need precise, quantitative analysis of multiple PTMs on histone tails. Both methods can be complementary.
Is it possible to analyse multiple histone PTMs simultaneously?
Absolutely. Modern mass spectrometry platforms allow simultaneous detection and quantification of dozens of histone marks from small sample amounts, enabling combinatorial PTM profiling.
What sample types can be used for histone PTM analysis?
Histone PTMs can be analysed from fresh or frozen tissues, cell lines, formalin-fixed paraffin-embedded (FFPE) samples, and even circulating tumor cells (CTCs) in some workflows.
References
- Audia J E, Campbell R M. Histone modifications and cancer. Cold Spring Harbor perspectives in biology, 2016, 8(4): a019521. DOI: 10.1101/cshperspect.a019521
- Xu X, et al. Metabolic reprogramming and epigenetic modifications in cancer: from the impacts and mechanisms to the treatment potential. Experimental & molecular medicine, 2023, 55(7): 1357-1370. DOI: 10.1038/s12276-023-01020-1
- Zhuang J, et al. Perspectives on the role of histone modification in breast cancer progression and the advanced technological tools to study epigenetic determinants of metastasis. Frontiers in genetics, 2020, 11: 603552. DOI: 10.3389/fgene.2020.603552
Our products and services are for research use only.