Mechanisms of Lactylation in Biological Systems
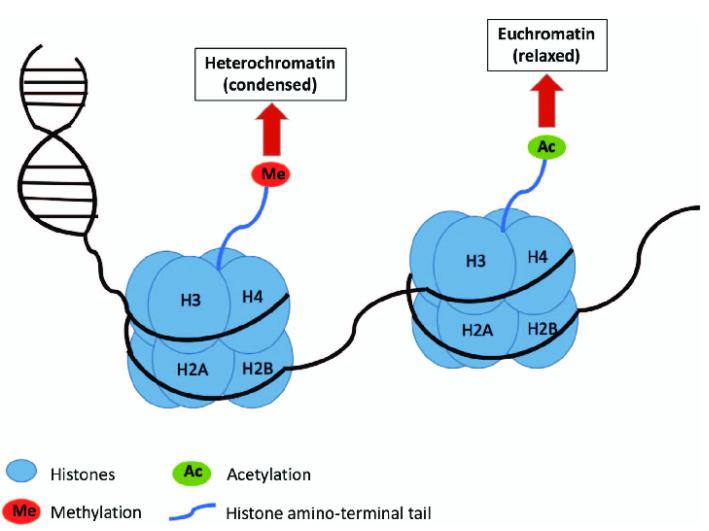
Lactylation represents a complex and nuanced mechanism within biological systems. Specifically, lactylation of histones exerts its regulatory effects through epigenetic pathways, while non-histone lactylation finely modulates protein function through various mechanisms, including steric hindrance, conformational changes, and charge neutralization. Such regulatory processes ultimately influence critical characteristics of proteins, including molecular interactions, stability, subcellular localization, and enzymatic activity.
Upon successfully identifying and characterizing differential lactylation sites relevant to the research, the focus shifts to further elucidating the underlying mechanisms. The role of histone lactylation in gene expression regulation is relatively well-defined, and subsequent research directions are clear; thus, this aspect will not be elaborated upon herein. Conversely, the mechanisms governing non-histone lactylation are notably diverse and complex.
To explore this field comprehensively, this article presents specific case studies of non-histone lactylation in the regulation of protein function, providing valuable insights and considerations for investigating the mechanisms underlying non-histone lactylation. Particular emphasis will be placed on how lactylation regulates protein interactions with other molecular entities, a critical component in understanding the overall mechanistic implications of non-histone lactylation.
Case Study on Lactylation in Protein-Molecule Interactions
Lactylation and Chemoresistance:
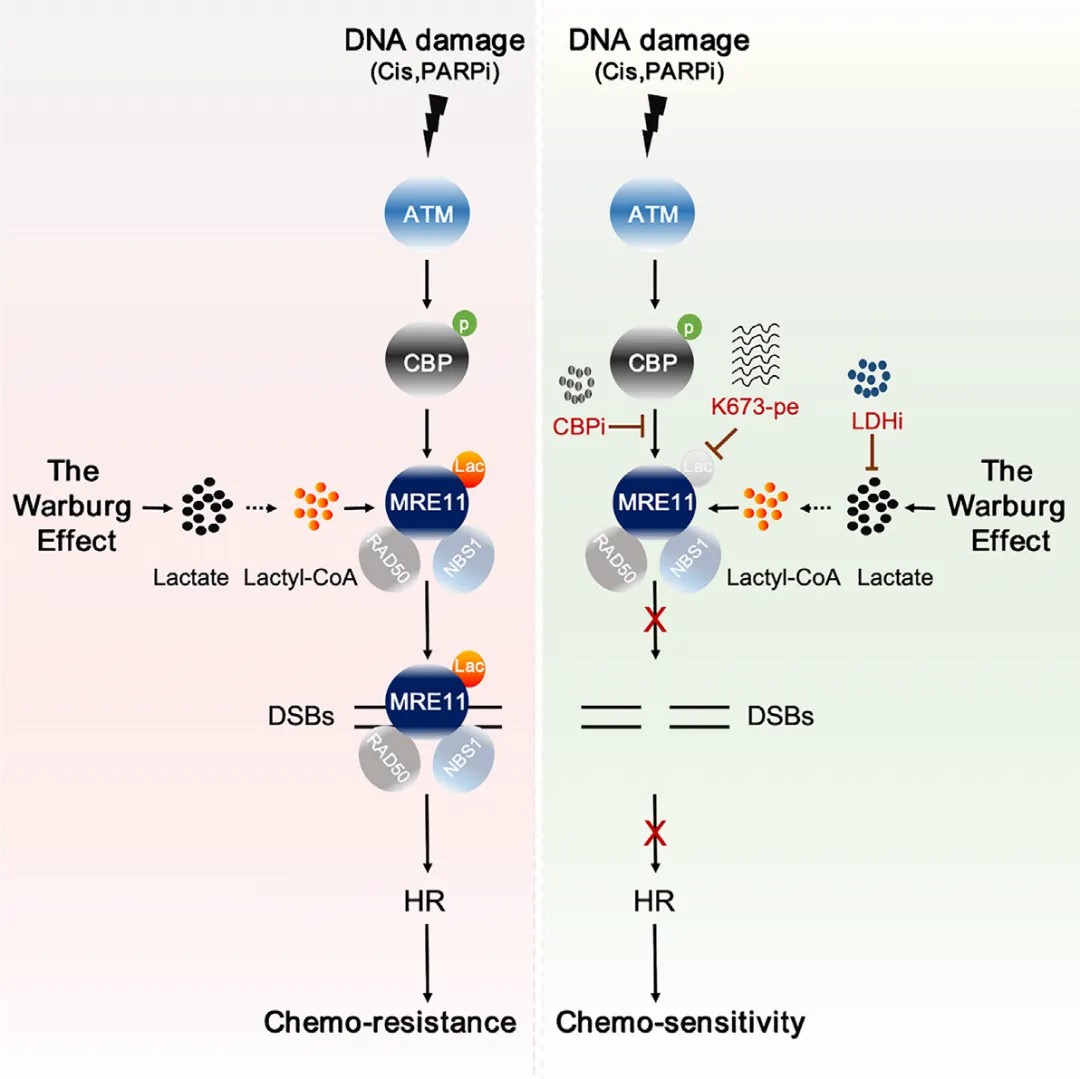
Metabolic Regulation of Homologous Recombination Repair by MRE11 Lactylation
This study conclusively demonstrates that MRE11, a critical protein involved in homologous recombination (HR), undergoes lactylation at lysine 678 (K678) by the CBP acetyltransferase, exhibiting significant functionality in response to DNA damage. The lactylation of MRE11 substantially enhances its binding affinity to DNA, thereby accelerating DNA end resection and the HR process. Inhibition of CBP or lactate dehydrogenase (LDH) results in downregulation of MRE11 lactylation, impeding HR and significantly increasing the chemosensitivity of tumor cells in patient-derived xenograft and organoid models. A specially designed cell-penetrating peptide, which selectively blocks the lactylation of MRE11, effectively inhibits HR and markedly enhances cancer cell sensitivity to cisplatin and PARP inhibitors (PARPi).
These findings not only reveal lactylation as a pivotal regulator of HR but also provide novel insights into the intricate relationship between cellular metabolism and double-strand break (DSB) repair. Furthermore, the results suggest that the Warburg effect may confer chemoresistance through enhanced HR pathways, proposing a potential therapeutic strategy aimed at targeting MRE11 lactylation to mitigate this adverse effect.
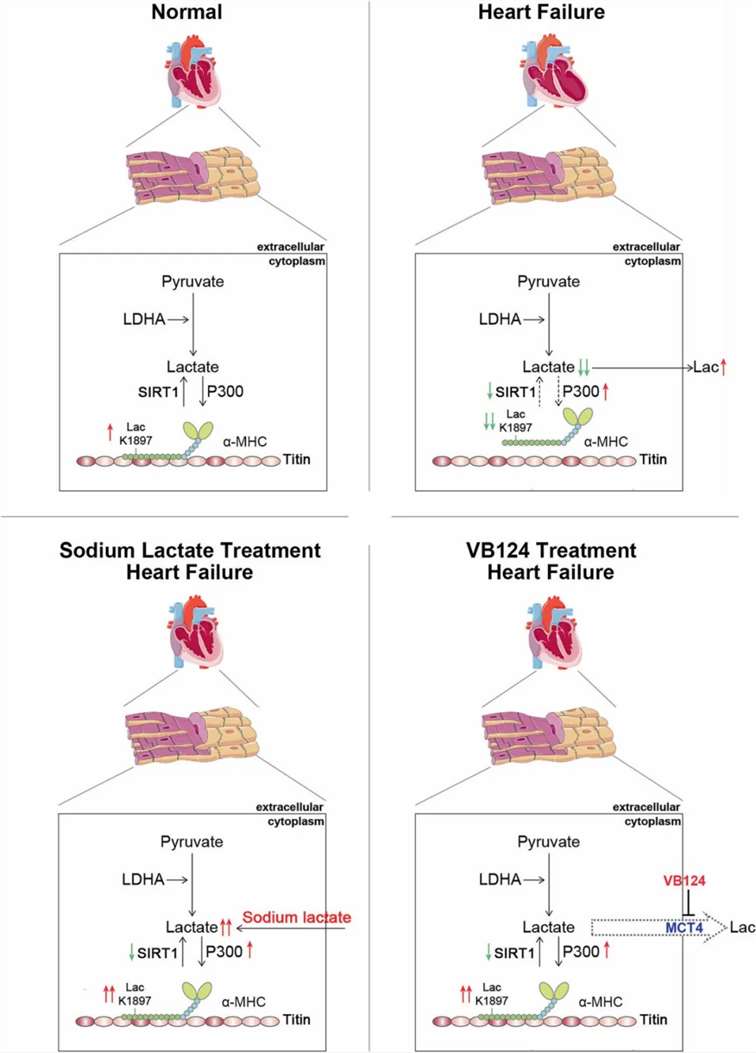
Lactylation and Heart Failure:
a-myosin heavy chain lactylation maintains sarcomericstructure and function and alleviates the development of heart failure
The interaction of α-myosin heavy chain (α-MHC) with Titin is essential for cardiac structure and contraction. However, the regulatory mechanisms of this interaction in both healthy and failing hearts remain inadequately understood. Lactate serves as a critical energy substrate in cardiac tissue. This study identifies lactylation of α-MHC at lysine 1897 (K1897) as a regulator of its interaction with Titin. A reduction in α-MHC K1897 lactylation was observed in both mice and patients with heart failure. In α-MHC K1897R knock-in mice, the absence of K1897 lactylation diminishes the α-MHC–Titin interaction, leading to impaired cardiac structure and function. Furthermore, p300 and Sirtuin 1 function as the acyltransferase and delactylase for α-MHC, respectively. Decreased lactate production, via chemical or genetic intervention, lowers α-MHC lactylation, disrupts α-MHC–Titin interaction, and exacerbates heart failure. Conversely, increasing lactate levels through sodium lactate administration or inhibiting key lactate transporters in cardiomyocytes enhances α-MHC K1897 lactylation and the α-MHC–Titin interaction, alleviating heart failure. Thus, α-MHC lactylation is dynamically regulated and serves as a crucial determinant of cardiac structure and function. Excessive lactate efflux and consumption by cardiomyocytes lead to reduced intracellular lactate levels, which is primarily responsible for diminished α-MHC K1897 lactylation during myocardial injury. This study elucidates the direct modulation of sarcomeric structure and function by cardiac metabolism through lactate-dependent modifications of α-MHC.
Introduction
A sarcomere represents the fundamental unit of cardiac structure and contraction. Within the sarcomere, the myosin tail interacts with Titin to form thick filaments, while the myosin head and neck domains establish cross-bridges. In the presence of calcium ions, the myosin head binds adenosine triphosphate (ATP), undergoes a conformational change, and interacts with thin filaments, resulting in cardiac contraction. Recent advancements in cryo-electron microscopy have illuminated this structure. Despite the known dynamic interactions between myosin and Titin, the mechanisms governing their association and dissociation remain elusive.
Traditionally viewed as a metabolic byproduct, lactate is now recognized as a vital energy substrate for cardiac function. It serves as a critical source for mitochondrial respiration and acts as an intercellular signaling molecule, thereby influencing metabolic regulation in vivo. Increasing evidence underscores lactate's role in cardiac hypertrophy, injury, and heart failure. For instance, lactate has been shown to be essential for hypertrophic growth in response to pressure overload, stabilizing proteins such as NDRG3 and stimulating extracellular signal-regulated kinase pathways. The absence of lactate can hinder hypertrophy development, exacerbating heart failure under pressure overload conditions. Furthermore, studies indicate that myocardial lactate levels significantly decrease following adriamycin-induced injury, leading to an increase in lactate secretion from cardiomyocytes, which correlates with adverse cardiovascular outcomes in heart failure patients. Interventions that inhibit lactate efflux from cardiomyocytes, such as targeting monocarboxylate transporter 4 (MCT4), demonstrate the protective role of intracellular lactate in alleviating heart failure. Notably, lactate consumption nearly doubles in failing hearts, and treatments with lactate analogs have shown promise in mitigating acute heart failure in animal models. Clinical evidence further supports these findings, revealing that sodium lactate administration can restore cardiac function in acute heart failure patients.
Recent investigations have also revealed that lactate can exert biological effects through protein lactylation, a post-translational modification that alters the function of both histone and non-histone proteins by modifying specific lysine residues. Examples include the lactylation of H3K18, which influences macrophage polarization, and the inhibition of adenylate kinase 2 in hepatocellular carcinoma via K28 lactylation, promoting tumor proliferation. Despite the acknowledged importance of lactate as an energy substrate, the physiological and pathological roles of lactylation in cardiomyocytes remain poorly characterized.
In this study, it is confirmed that lactylation of α-MHC at lysine 1897 is diminished in both mice and patients with heart failure. The α-MHC K1897R knock-in (KI) mice exhibit impaired α-MHC–Titin interaction and more severe heart failure. Additionally, p300 and Sirtuin 1 are identified as the acyltransferase and delactylase for α-MHC, respectively. Notably, excessive lactate efflux and consumption by cardiomyocytes lead to decreased intracellular lactate levels, which is primarily responsible for reduced α-MHC K1897 lactylation and α-MHC–Titin interaction in myocardial injury. Treatment with sodium lactate or inhibition of lactate efflux can restore α-MHC K1897 lactylation and α-MHC–Titin interaction, offering a therapeutic approach to heart failure management. These findings underscore the critical role of endogenous lactate in regulating sarcomeric structure and function.
Lactylation and Tumor Therapeutic Resistance:
Glycometabolic reprogramming-induced XRCC1actylation confers therapeutic resistance inALDH1A3-overexpressing glioblastoma
Patients with high aldehyde dehydrogenase 1A3 (ALDH1A3) expressing glioblastoma (ALDH1A3^hi GBM) exhibit limited benefits from postoperative chemoradiotherapy. Therefore, elucidating the mechanisms of therapeutic resistance in these patients is crucial for advancing novel treatment strategies. This study focuses on the interaction between ALDH1A3 and pyruvate kinase M2 (PKM2), revealing that this interaction significantly enhances the tetramerization of PKM2, thereby promoting the accumulation of lactate in glioblastoma stem cells (GSCs).
Through meticulous analysis of the lactylated proteome in GSCs, a significant finding emerged: lactylation modification occurs at lysine 247 (K247) on X-ray cross-complementing protein 1 (XRCC1). This modification substantially increases the affinity of XRCC1 for importin α, facilitating the nuclear translocation of XRCC1 and ultimately enhancing DNA repair processes.
Building upon these findings, a small-molecule compound, D34-919, was identified. This compound effectively disrupts the interaction between ALDH1A3 and PKM2, thereby inhibiting the ALDH1A3-mediated tetramerization of PKM2. Subsequent investigations demonstrated that D34-919 treatment significantly augments apoptosis in GBM cells induced by chemoradiotherapy, thereby offering a promising potential therapeutic strategy for patients with ALDH1A3^hi GBM.
In summary, this research elucidates a pivotal mechanism by which ALDH1A3 overexpression induces therapeutic resistance through the lactylation of XRCC1, emphasizing the relevance of glycometabolic reprogramming in glioblastoma treatment resistance and suggesting novel avenues for intervention.
Protein-Protein Interactions:
Integrated Lactylome Characterization Reveals the Molecular Dynamics of Protein Regulation in Gastrointestinal Cancers.
Patients diagnosed with gastrointestinal (GI) cancers exhibit complex molecular alterations, underscoring the need for comprehensive biomolecular profiling to inform treatment strategies. This study presents a global lactylome characterization across various GI cancer types, identifying over 11,000 lysine lactylation (Kla) sites in malignant tissues from a cohort of 40 patients.
The integrated lactylome analysis reveals that Kla significantly modulates proteins involved in critical cancer processes, such as epigenetic reprogramming and metabolic dysregulation. Notably, 37 Kla sites were consistently upregulated across multiple GI cancers, implicating their role in gene regulation. Among these, lactylation of chromobox 3 (CBX3) at lysine 10 (K10) was shown to enhance its interaction with the epigenetic marker H3K9me3, thereby promoting tumor progression.
Building on these findings, this research underscores the potential of lactylation modifications as therapeutic targets. The identification of specific Kla sites associated with oncogenic processes provides new insights into the molecular dynamics governing GI cancers. Overall, this study establishes a foundational understanding of the lactylome landscape in GI malignancies, paving the way for future drug discovery and novel therapeutic interventions.
Protein-RNA interactions:
Lactylation-driven METTL3-mediated RNA m6A modification promotes immunosuppression of tumor-infiltrating myeloid cells
This study explores the impact of lactylation-driven METTL3-mediated RNA N6-methyladenosine (m6A) modification on the immunosuppressive behavior of tumor-infiltrating myeloid cells (TIMs). It establishes a significant link between high METTL3 expression in TIMs and adverse outcomes in colon cancer patients. The research demonstrates that lactate present in the tumor microenvironment (TME) stimulates the upregulation of METTL3 through H3K18 lactylation, leading to enhanced m6A modification of Jak1 mRNA, which in turn boosts JAK1 protein translation and activates the STAT3 pathway.
Key findings highlight specific lactylation sites within METTL3's zinc-finger domain, essential for its capacity to bind RNA. This lactylation-METTL3-m6A-JAK/STAT3 signaling axis strengthens the immunosuppressive functions of myeloid cells, playing a crucial role in tumor immune evasion. The research underscores the potential of targeting this pathway as a novel therapeutic approach in colorectal cancer, emphasizing the complex interactions between lactate, METTL3, and immunosuppressive mechanisms within the TME.
In summary, this investigation reveals vital molecular dynamics that drive the immunosuppressive properties of TIMs and provides valuable insights into strategies aimed at boosting antitumor immunity.
Summary of Molecular Interactions Regulated by Lactylation
The molecular interactions involving lactylation-regulated proteins encompass protein-protein, protein-DNA, and protein-RNA interactions. The focal point of research is determined by the specific function of the lactylated proteins. If the function of a protein has not been definitively linked to its binding with DNA or RNA, priority should be given to investigating the effects of lactylation on protein-protein interactions. Conversely, if it is established that a protein exerts its function through interactions with DNA or RNA, the analysis of protein-DNA/RNA interactions should be prioritized.
In the validation process, methodologies may be employed to create conditions of high lactylation (e.g., treatment with lactate or sodium lactate) or low lactylation (e.g., K→R site mutations, inhibition, or knockout of lactate-producing enzymes). These experimental conditions enable the assessment of binding strength between the target lactylated proteins and their associated molecules under varying lactylation states, thereby clarifying whether lactylation enhances or diminishes specific protein-molecule interactions.
This systematic approach facilitates a comprehensive understanding of the functional implications of lactylation in cellular processes and provides insights into the regulatory mechanisms governing protein interactions within biological systems.
Our products and services are for research use only.