
- Home
- PTMs Proteomics
- Modificated PROTACs Proteomics Service
Proteolysis targeting chimeras (PROTACs) is a chemical knockout strategy that has emerged as a promising technique for regulating proteins of interest (POIs) through degradation.
Small-molecule inhibitor (SMI) development of disease-associated proteins has many limitations: lack of suitable binding pockets to directly modulate protein function; often requires high exposure to achieve sufficient site occupancy resulting in risk of off-target toxicity; poor selectivity against active sites of proteins such as kinases.
The strategy of targeting specific proteins for post-translational degradation is the key to circumvent the above defects. Most proteins undergo degradation via the ubiquitin-proteasome system (UPS), in which proteins are first covalently labeled with ubiquitin and then sent to the 26S proteasome for degradation. PROTAC technology utilizes UPS to degrade specific proteins (POIs) (Figure 1).
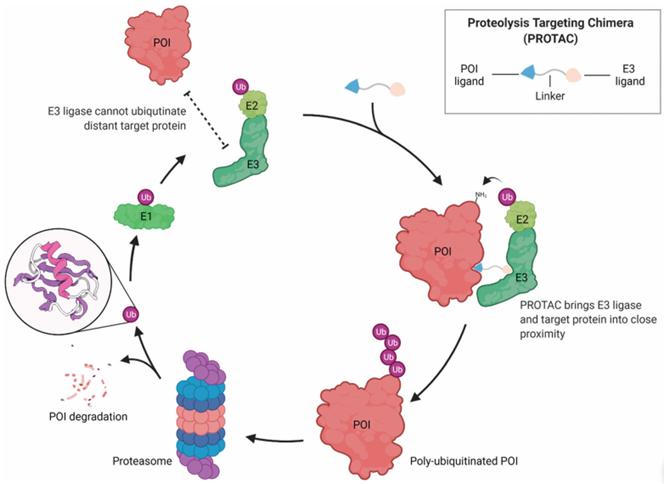 Figure 1 PROTACs use the UPS to induce targeted protein degradation
Figure 1 PROTACs use the UPS to induce targeted protein degradation
In terms of molecular structure, the PROTACs drug molecule consists of three parts: the target protein ligand, the E3 ubiquitinated ligase, and the Linker that connects the two. PROTACs technology looks like dumbbells. This bifunctional molecule connects the ligand of the protein of interest as well as the recruiting ligand of the E3 ubiquitin ligase through an ideal linker. That is, the PROTACs technology binds to the protein of interest (i.e., the target protein) at one end and the E3 ubiquitin ligase at the other. E3 ubiquitin ligases mark proteins of interest as defective or damaged by attaching a small protein called ubiquitin to them. The tagged protein of interest is then disposed of by the cell's protein shredder, the proteasome. That is, degradation occurs when PROTACs promote the formation of a ternary complex between the protein of interest and the E3 ubiquitin ligase, thereby regulating the level of the protein of interest.
One application of PROTACs is to induce ubiquitination of overdose of target proteins when administered, thereby limiting the side effects of excess drug. PROTAC proteomics is a qualitative and quantitative analysis of the proteome after the action of PROTACs to confirm the degradation of the target protein and potential off-target situations; at the same time, it conducts quantitative analysis of the ubiquitination level to investigate the ubiquitination of the target protein.
Creative Roteomics has officially entered the field of pharmaceutical research and development with its PROTAC drug development service based on proteomics. This service offers the following advantages:
(1) Wide-ranging non-target protein detection based on DIA and TMT, analyzing the degradation efficiency of target proteins and off-target effects after PROTAC treatment.
(2) Targeted quantitative detection technology based on MRM/PRM, quantifying the degradation levels of target proteins after PROTAC treatment and calculating parameters such as DC50, DC90, or DCmax.
(3) Protein-protein interaction validation between target proteins and E3 ubiquitin ligases based on AP-MS interactome proteomics technology.
(4) Study of the ubiquitination mechanisms involved in the degradation of target proteins after PROTAC treatment based on ubiquitin proteomics technology.
(5) Protein quantification using Simple/Digital Western (an upgraded version of traditional Western blot), offering accurate and highly reproducible results with high detection efficiency. It allows assessment of target protein degradation at the cellular level, organ models, animals, and other different levels, determining dose-response relationships and calculating parameters such as DC50 or DC90. It serves as a powerful tool for PROTAC development.
Sample Requirement
protein extraction, peptide preparation → bRPLC fractionation, online LC-MS/MS → bioinformatics analysis
Protein qualitative and quantitative results
1) Qualitative analysis of peptides and corresponding proteins, quantification of differentially expressed peptides and corresponding proteins
2) Visual view presentation: Venn diagram, volcano diagram, cluster heat map, GO (subcellular localization, molecular function, biological process), KEGG signaling pathway, WGCNA.
Our products and services are for research use only.